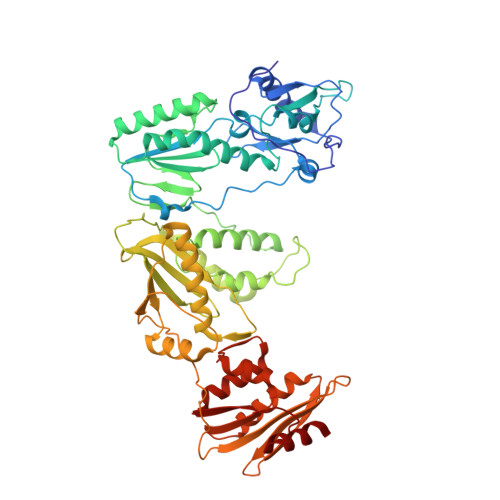
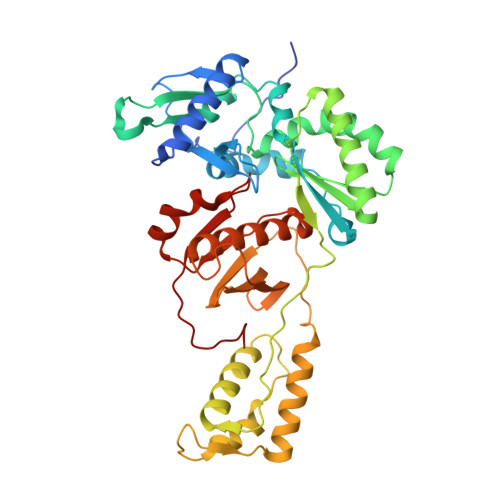
Structural Insights into HIV Reverse Transcriptase Mutations Q151M and Q151M Complex That Confer Multinucleoside Drug Resistance.
Das, K., Martinez, S.E., Arnold, E.(2017) Antimicrob Agents Chemother 61
- PubMed: 28396546
- DOI: https://doi.org/10.1128/AAC.00224-17
- Primary Citation of Related Structures:
5TXL, 5TXM, 5TXN, 5TXO, 5TXP - PubMed Abstract:
HIV-1 reverse transcriptase (RT) is targeted by multiple drugs. RT mutations that confer resistance to nucleoside RT inhibitors (NRTIs) emerge during clinical use. Q151M and four associated mutations, A62V, V75I, F77L, and F116Y, were detected in patients failing therapies with dideoxynucleosides (didanosine [ddI], zalcitabine [ddC]) and/or zidovudine (AZT). The cluster of the five mutations is referred to as the Q151M complex (Q151Mc), and an RT or virus containing Q151Mc exhibits resistance to multiple NRTIs. To understand the structural basis for Q151M and Q151Mc resistance, we systematically determined the crystal structures of the wild-type RT/double-stranded DNA (dsDNA)/dATP (complex I), wild-type RT/dsDNA/ddATP (complex II), Q151M RT/dsDNA/dATP (complex III), Q151Mc RT/dsDNA/dATP (complex IV), and Q151Mc RT/dsDNA/ddATP (complex V) ternary complexes. The structures revealed that the deoxyribose rings of dATP and ddATP have 3'-endo and 3'-exo conformations, respectively. The single mutation Q151M introduces conformational perturbation at the deoxynucleoside triphosphate (dNTP)-binding pocket, and the mutated pocket may exist in multiple conformations. The compensatory set of mutations in Q151Mc, particularly F116Y, restricts the side chain flexibility of M151 and helps restore the DNA polymerization efficiency of the enzyme. The altered dNTP-binding pocket in Q151Mc RT has the Q151-R72 hydrogen bond removed and has a switched conformation for the key conserved residue R72 compared to that in wild-type RT. On the basis of a modeled structure of hepatitis B virus (HBV) polymerase, the residues R72, Y116, M151, and M184 in Q151Mc HIV-1 RT are conserved in wild-type HBV polymerase as residues R41, Y89, M171, and M204, respectively; functionally, both Q151Mc HIV-1 and wild-type HBV are resistant to dideoxynucleoside analogs.
Organizational Affiliation:
Center for Advanced Biotechnology and Medicine and Department of Chemistry and Chemical Biology, Rutgers University, Piscataway, New Jersey, USA.