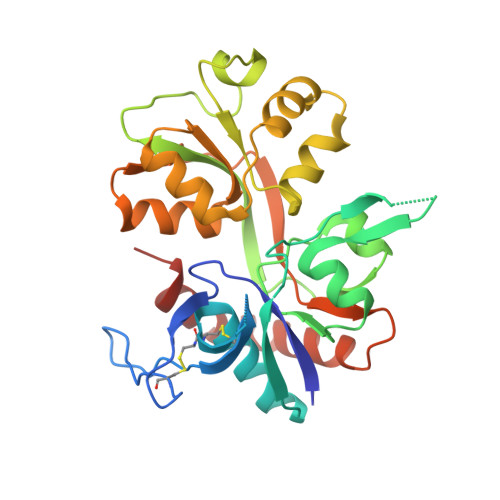
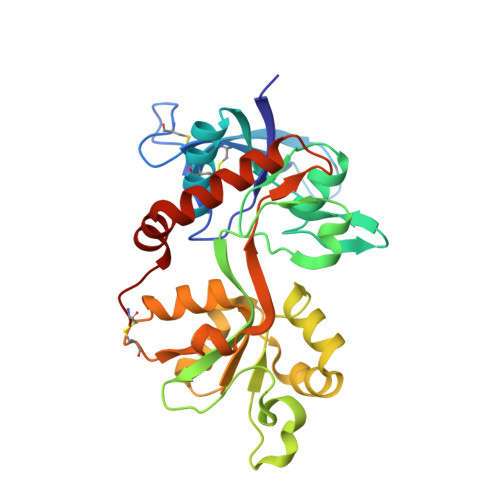
Structural basis of subunit selectivity for competitive NMDA receptor antagonists with preference for GluN2A over GluN2B subunits.
Lind, G.E., Mou, T.C., Tamborini, L., Pomper, M.G., De Micheli, C., Conti, P., Pinto, A., Hansen, K.B.(2017) Proc Natl Acad Sci U S A 114: E6942-E6951
- PubMed: 28760974
- DOI: https://doi.org/10.1073/pnas.1707752114
- Primary Citation of Related Structures:
5DEX, 5VIH, 5VII, 5VIJ - PubMed Abstract:
NMDA-type glutamate receptors are ligand-gated ion channels that contribute to excitatory neurotransmission in the central nervous system (CNS). Most NMDA receptors comprise two glycine-binding GluN1 and two glutamate-binding GluN2 subunits (GluN2A-D). We describe highly potent ( S )-5-[( R )-2-amino-2-carboxyethyl]-4,5-dihydro-1 H -pyrazole-3-carboxylic acid (ACEPC) competitive GluN2 antagonists, of which ST3 has a binding affinity of 52 nM at GluN1/2A and 782 nM at GluN1/2B receptors. This 15-fold preference of ST3 for GluN1/2A over GluN1/2B is improved compared with NVP-AAM077, a widely used GluN2A-selective antagonist, which we show has 11-fold preference for GluN1/2A over GluN1/2B. Crystal structures of the GluN1/2A agonist binding domain (ABD) heterodimer with bound ACEPC antagonists reveal a binding mode in which the ligands occupy a cavity that extends toward the subunit interface between GluN1 and GluN2A ABDs. Mutational analyses show that the GluN2A preference of ST3 is primarily mediated by four nonconserved residues that are not directly contacting the ligand, but positioned within 12 Å of the glutamate binding site. Two of these residues influence the cavity occupied by ST3 in a manner that results in favorable binding to GluN2A, but occludes binding to GluN2B. Thus, we reveal opportunities for the design of subunit-selective competitive NMDA receptor antagonists by identifying a cavity for ligand binding in which variations exist between GluN2A and GluN2B subunits. This structural insight suggests that subunit selectivity of glutamate-site antagonists can be mediated by mechanisms in addition to direct contributions of contact residues to binding affinity.
Organizational Affiliation:
Department of Biomedical and Pharmaceutical Sciences, University of Montana, Missoula, MT 59812.