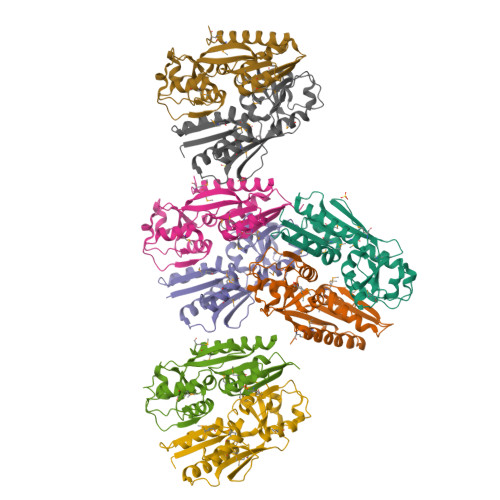
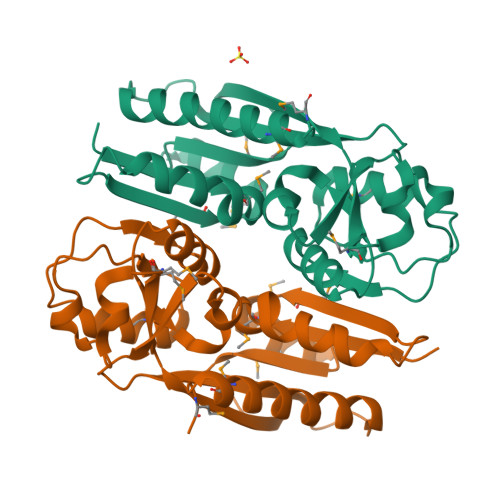
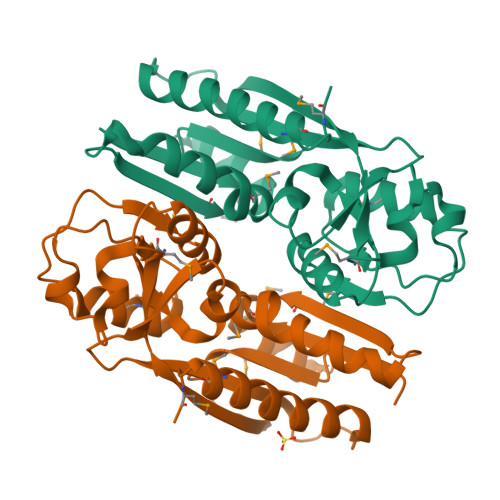
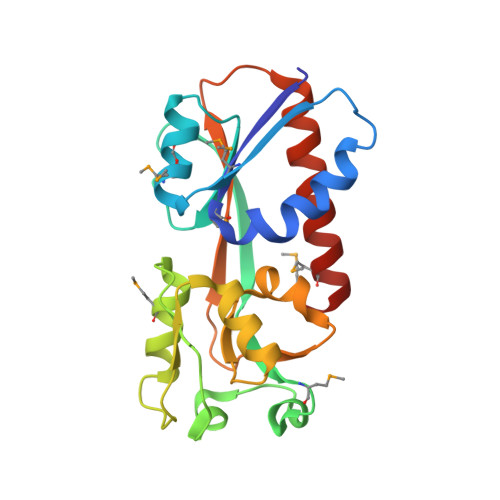
Crystal Structure of the Ligand-Binding Domain of a LysR-type Transcriptional Regulator: Transcriptional Activation via a Rotary Switch.
Kim, Y., Chhor, G., Tsai, C.S., Winans, J.B., Jedrzejczak, R., Joachimiak, A., Winans, S.C.(2018) Mol Microbiol
- PubMed: 30168204
- DOI: https://doi.org/10.1111/mmi.14115
- Primary Citation of Related Structures:
5VVH, 5VVI - PubMed Abstract:
LysR-type transcriptional regulators (LTTRs) generally bind to target promoters in two conformations, depending on the availability of inducing ligands. OccR is an LTTR that regulates the octopine catabolism operon of Agrobacterium tumefaciens. OccR binds to a site located between the divergent occQ and occR promoters. Octopine triggers a conformational change that activates the occQ promoter, and does not affect autorepression. This change shortens the length of bound DNA and relaxes a high-angle DNA bend. Here, we describe the crystal structure of the ligand-binding domain (LBD) of OccR apoprotein and holoprotein. Pairs of LBDs form dimers with extensive hydrogen bonding, while pairs of dimers interact via a single helix, creating a tetramer interface. Octopine causes a 70° rotation of each dimer with respect to the opposite dimer, precisely at the tetramer interface. We modeled the DNA binding domain (DBD), linker helix and bound DNA onto the apoprotein and holoprotein. The two DBDs of the modeled apoprotein lie far apart and the bound DNA between them has a high-angle DNA bend. In contrast, the two DBDs of the holoprotein lie closer to each other, with a low DNA bend angle. This inter-dimer pivot fully explains earlier studies of this LTTR.
Organizational Affiliation:
Midwest Center for Structural Genomics, Biosciences, Argonne National Laboratory, Argonne, IL, 60439, USA.