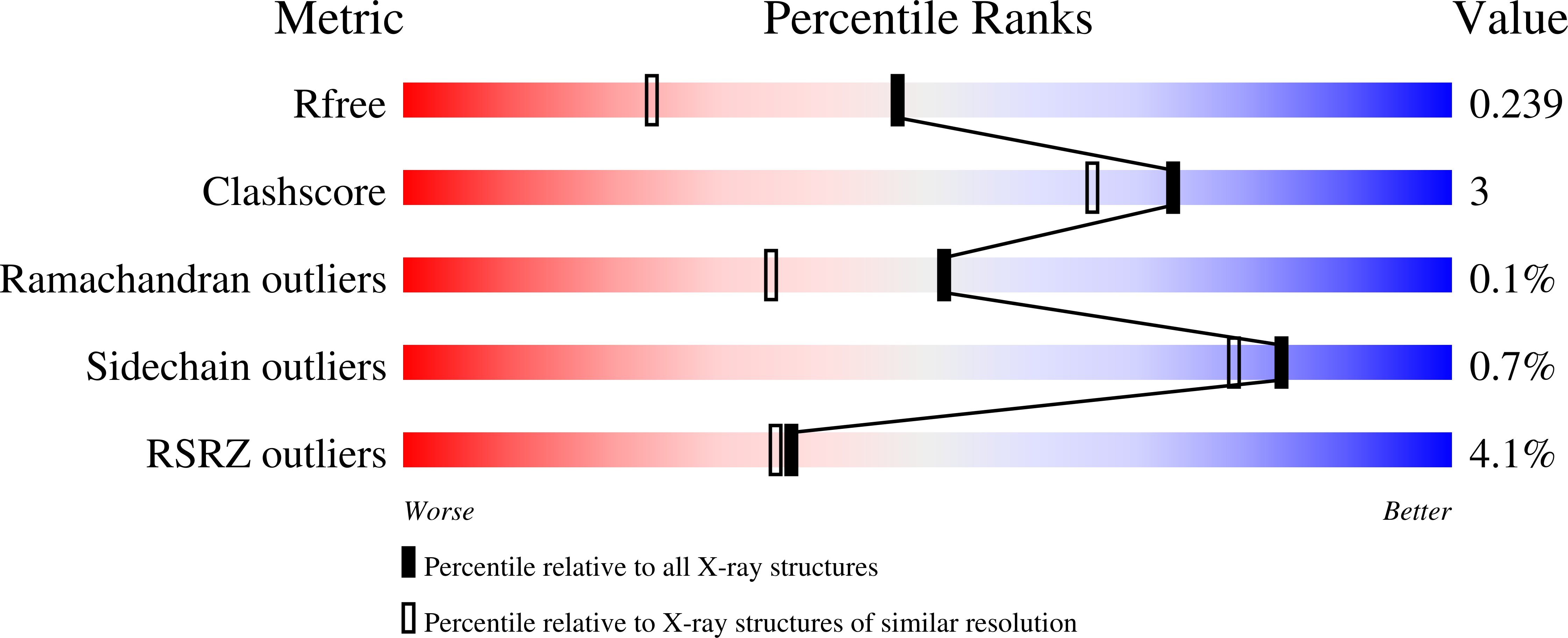
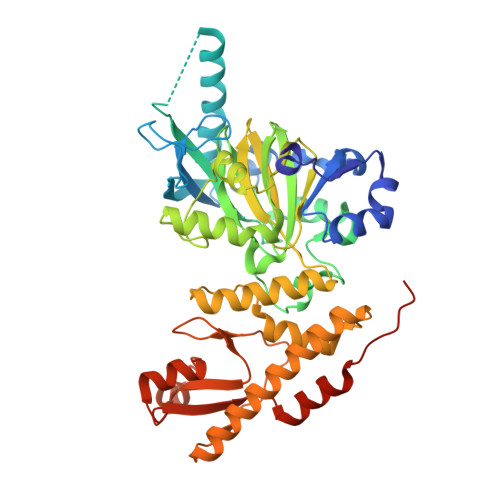
Crystal Structure of Jmjc Domain of Human Histone Demethylase Uty in Complex with N08619B
Nowak, R., Krojer, T., Johansson, C., Gileadi, C., Kupinska, K., Pearce, N.M., von Delft, F., Arrowsmith, C.H., Bountra, C., Edwards, A., Oppermann, U.To be published.