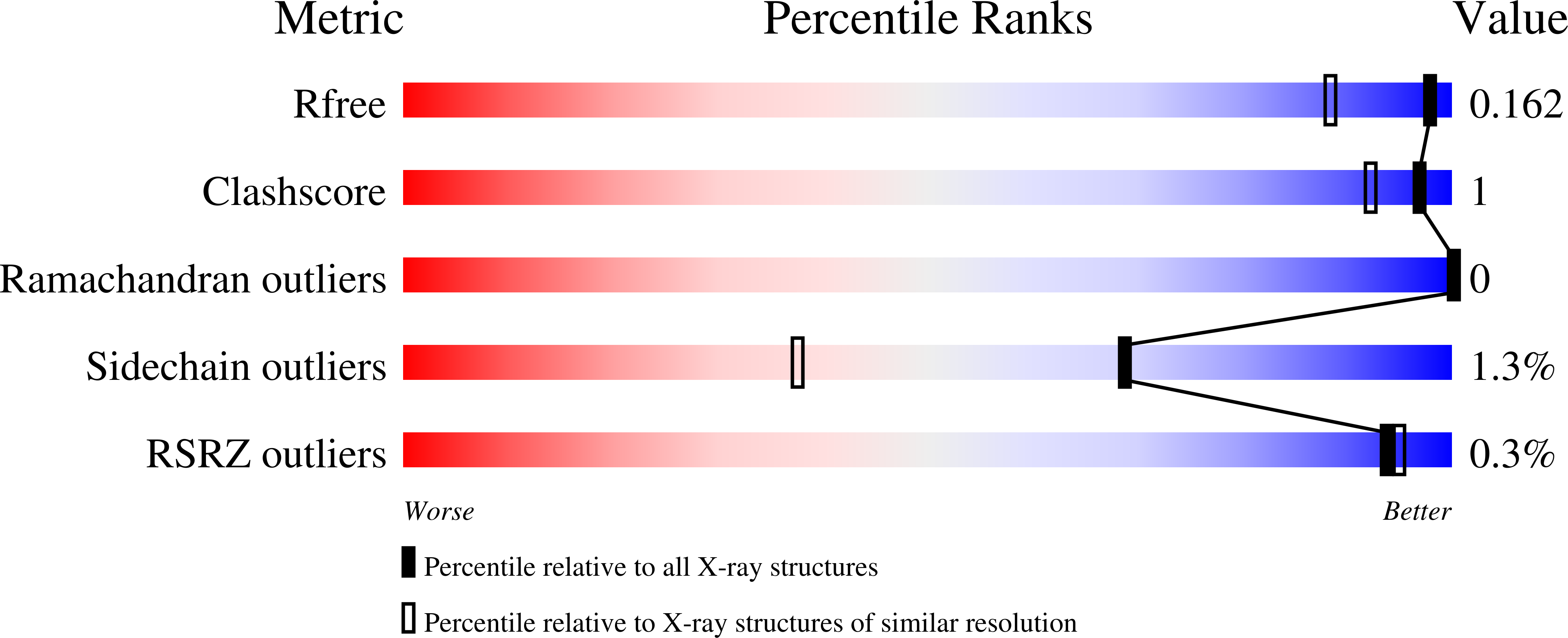
Control and regulation of S-Adenosylmethionine biosynthesis by the regulatory beta subunit and quinolone-based compounds.
Panmanee, J., Bradley-Clarke, J., Mato, J.M., O'Neill, P.M., Antonyuk, S.V., Hasnain, S.S.(2019) FEBS J 286: 2135-2154
- PubMed: 30776190
- DOI: https://doi.org/10.1111/febs.14790
- Primary Citation of Related Structures:
6FBN, 6FBO, 6FBP, 6FCB, 6FCD, 6G6R - PubMed Abstract:
Methylation is an underpinning process of life and provides control for biological processes such as DNA synthesis, cell growth, and apoptosis. Methionine adenosyltransferases (MAT) produce the cellular methyl donor, S-Adenosylmethionine (SAMe). Dysregulation of SAMe level is a relevant event in many diseases, including cancers such as hepatocellular carcinoma and colon cancer. In addition, mutation of Arg264 in MATα1 causes isolated persistent hypermethioninemia, which is characterized by low activity of the enzyme in liver and high level of plasma methionine. In mammals, MATα1/α2 and MATβV1/V2 are the catalytic and the major form of regulatory subunits, respectively. A gating loop comprising residues 113-131 is located beside the active site of catalytic subunits (MATα1/α2) and provides controlled access to the active site. Here, we provide evidence of how the gating loop facilitates the catalysis and define some of the key elements that control the catalytic efficiency. Mutation of several residues of MATα2 including Gln113, Ser114, and Arg264 lead to partial or total loss of enzymatic activity, demonstrating their critical role in catalysis. The enzymatic activity of the mutated enzymes is restored to varying degrees upon complex formation with MATβV1 or MATβV2, endorsing its role as an allosteric regulator of MATα2 in response to the levels of methionine or SAMe. Finally, the protein-protein interacting surface formed in MATα2:MATβ complexes is explored to demonstrate that several quinolone-based compounds modulate the activity of MATα2 and its mutants, providing a rational for chemical design/intervention responsive to the level of SAMe in the cellular environment. ENZYMES: Methionine adenosyltransferase (EC.2.5.1.6). DATABASE: Structural data are available in the RCSB PDB database under the PDB ID 6FBN (Q113A), 6FBP (S114A: P22 1 2 1 ), 6FBO (S114A: I222), 6FCB (P115G), 6FCD (R264A), 6FAJ (wtMATα2: apo), 6G6R (wtMATα2: holo).
Organizational Affiliation:
Molecular Biophysics Group, Institute of Integrative Biology, Faculty of Health and Life Sciences, University of Liverpool, UK.