Isoxazolopyrimidine-Based Inhibitors ofPlasmodium falciparumDihydroorotate Dehydrogenase with Antimalarial Activity.
Kokkonda, S., El Mazouni, F., White, K.L., White, J., Shackleford, D.M., Lafuente-Monasterio, M.J., Rowland, P., Manjalanagara, K., Joseph, J.T., Garcia-Perez, A., Fernandez, J., Gamo, F.J., Waterson, D., Burrows, J.N., Palmer, M.J., Charman, S.A., Rathod, P.K., Phillips, M.A.(2018) ACS Omega 3: 9227-9240
- PubMed: 30197997
- DOI: https://doi.org/10.1021/acsomega.8b01573
- Primary Citation of Related Structures:
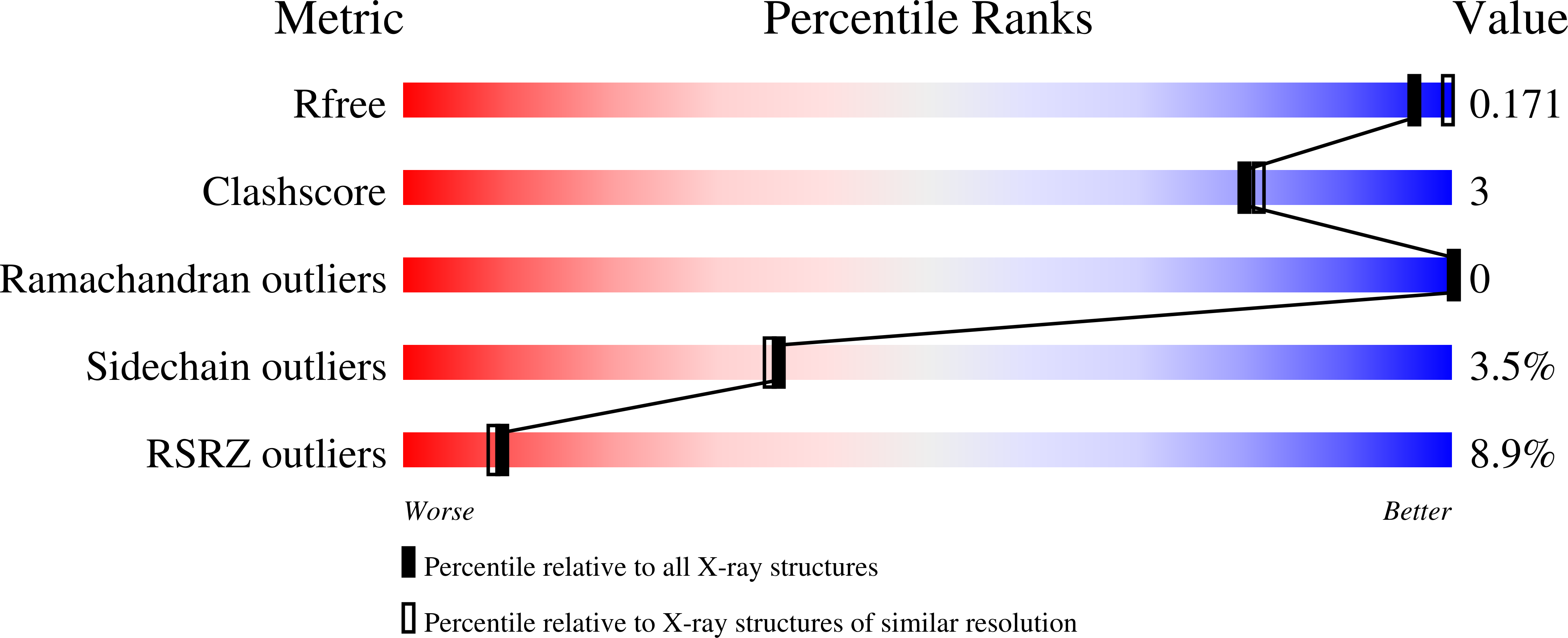
6GJG - PubMed Abstract:
Malaria kills nearly 0.5 million people yearly and impacts the lives of those living in over 90 countries where it is endemic. The current treatment programs are threatened by increasing drug resistance. Dihydroorotate dehydrogenase (DHODH) is now clinically validated as a target for antimalarial drug discovery as a triazolopyrimidine class inhibitor ( DSM265 ) is currently undergoing clinical development. We discovered a related isoxazolopyrimidine series in a phenotypic screen, later determining that it targeted DHODH. To determine if the isoxazolopyrimidines could yield a drug candidate, we initiated hit-to-lead medicinal chemistry. Several potent analogues were identified, including a compound that showed in vivo antimalarial activity. The isoxazolopyrimidines were more rapidly metabolized than their triazolopyrimidine counterparts, and the pharmacokinetic data were not consistent with the goal of a single-dose treatment for malaria.
Organizational Affiliation:
Departments of Chemistry and Global Health, University of Washington, Seattle, Washington 98195, United States.