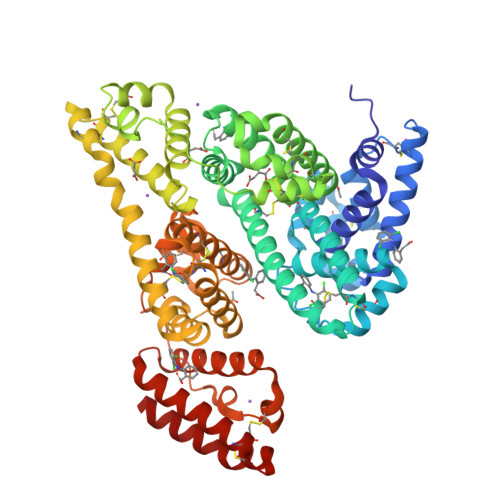
Structural Investigation of Diclofenac Binding to Ovine, Caprine, and Leporine Serum Albumins.
Talaj, J.A., Zielinski, K., Bujacz, A.(2023) Int J Mol Sci 24
- PubMed: 36675044
- DOI: https://doi.org/10.3390/ijms24021534
- Primary Citation of Related Structures:
6HN0, 6HN1, 8BSG - PubMed Abstract:
Free drug concentration in the blood sera is crucial for its appropriate activity. Serum albumin, the universal blood carrier protein, is responsible for transporting drugs and releasing them into the bloodstream. Therefore, a drug's binding to SA is especially important for its bioavailability and it is a key problem in the drug design process. In this paper, we present crystal structures of three animal serum albumin complexes: ovine, caprine, and leporine, with diclofenac, a popular non-steroidal anti-inflammatory drug that is used in therapy of chronic and acute pain. Details of diclofenac binding mode by the presented serum albumins are compared with analogous complexes of human and equine serum albumins. The analysis of the occupied binding pockets in crystal structures of the investigated serum albumins from different mammals shows that they have two common and a number of unique diclofenac binding sites. The most intriguing is the fact that the albumins from the described species are able to bind different numbers of molecules of this popular anti-inflammatory drug, but none of the binding sites overlap with ones in the human serum albumin.
Organizational Affiliation:
Department of Molecular Biology of Cancer, Medical University of Lodz, 6/8 Mazowiecka Street, 92-215 Lodz, Poland.