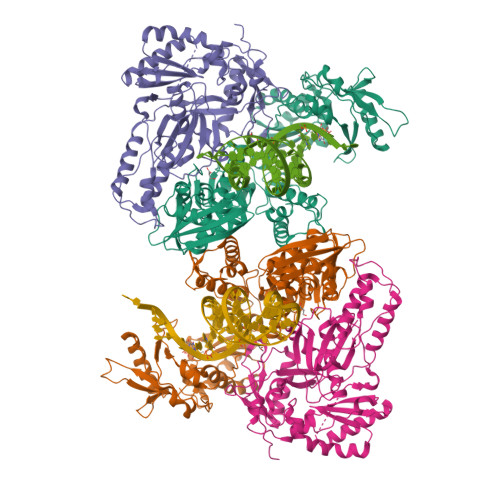
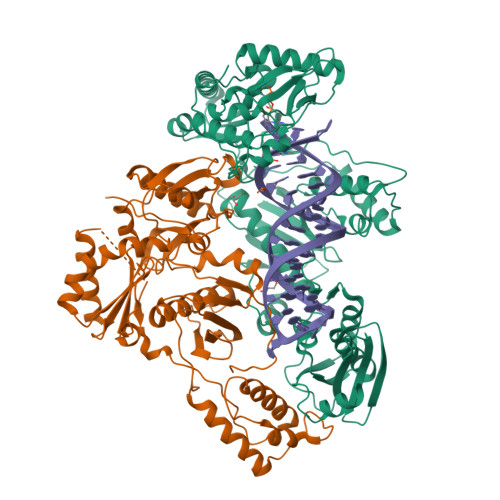
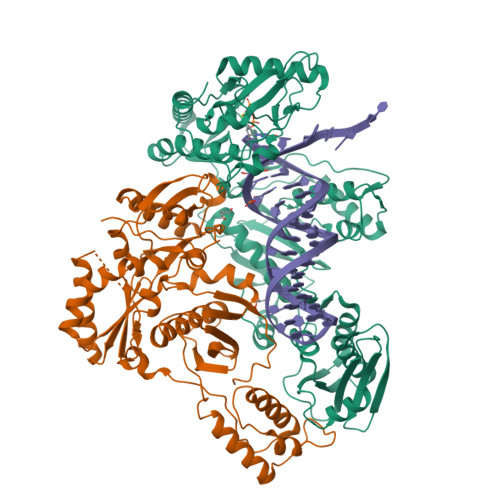
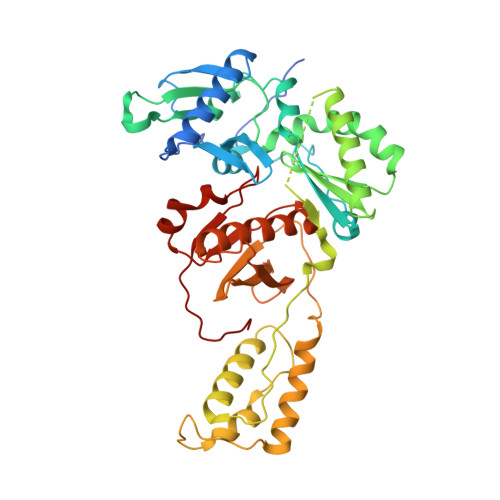
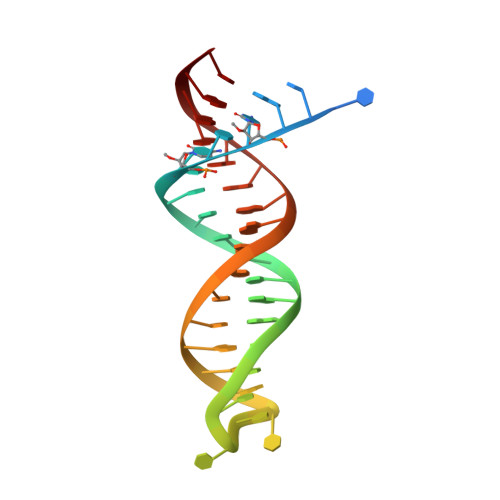
Active-site deformation in the structure of HIV-1 RT with HBV-associated septuple amino acid substitutions rationalizes the differential susceptibility of HIV-1 and HBV against 4'-modified nucleoside RT inhibitors.
Yasutake, Y., Hattori, S.I., Tamura, N., Matsuda, K., Kohgo, S., Maeda, K., Mitsuya, H.(2019) Biochem Biophys Res Commun 509: 943-948
- PubMed: 30648556
- DOI: https://doi.org/10.1016/j.bbrc.2019.01.026
- Primary Citation of Related Structures:
6IK9, 6IKA - PubMed Abstract:
Nucleoside analogue reverse transcriptase (RT) inhibitors (NRTIs) are major antiviral agents against hepatitis B virus (HBV) and human immunodeficiency virus type-1 (HIV-1). However, the notorious insoluble property of HBV RT has prevented atomic-resolution structural studies and rational anti-HBV drug design. Here, we created HIV-1 RT mutants containing HBV-mimicking sextuple or septuple amino acid substitutions at the nucleoside-binding site (N-site) and verified that these mutants retained the RT activity. The most active RT mutant, HIV-1 RT 7MC , carrying Q151M/G112S/D113A/Y115F/F116Y/F160L/I159L was successfully crystallized, and its three-dimensional structure was determined in complex with DNA:dGTP/entecavir-triphosphate (ETV-TP), a potent anti-HBV guanosine analogue RT inhibitor, at a resolution of 2.43 Å and 2.60 Å, respectively. The structures reveal significant positional rearrangements of the amino acid side-chains at the N-site, elucidating the mechanism underlying the differential susceptibility of HIV-1 and HBV against recently reported 4'-modified NRTIs.
Organizational Affiliation:
Bioproduction Research Institute, National Institute of Advanced Industrial Science and Technology (AIST), Sapporo, 062-8517, Japan; Computational Bio Big-Data Open Innovation Laboratory (CBBD-OIL), AIST, Sapporo, 062-8517, Japan. Electronic address: y-yasutake@aist.go.jp.