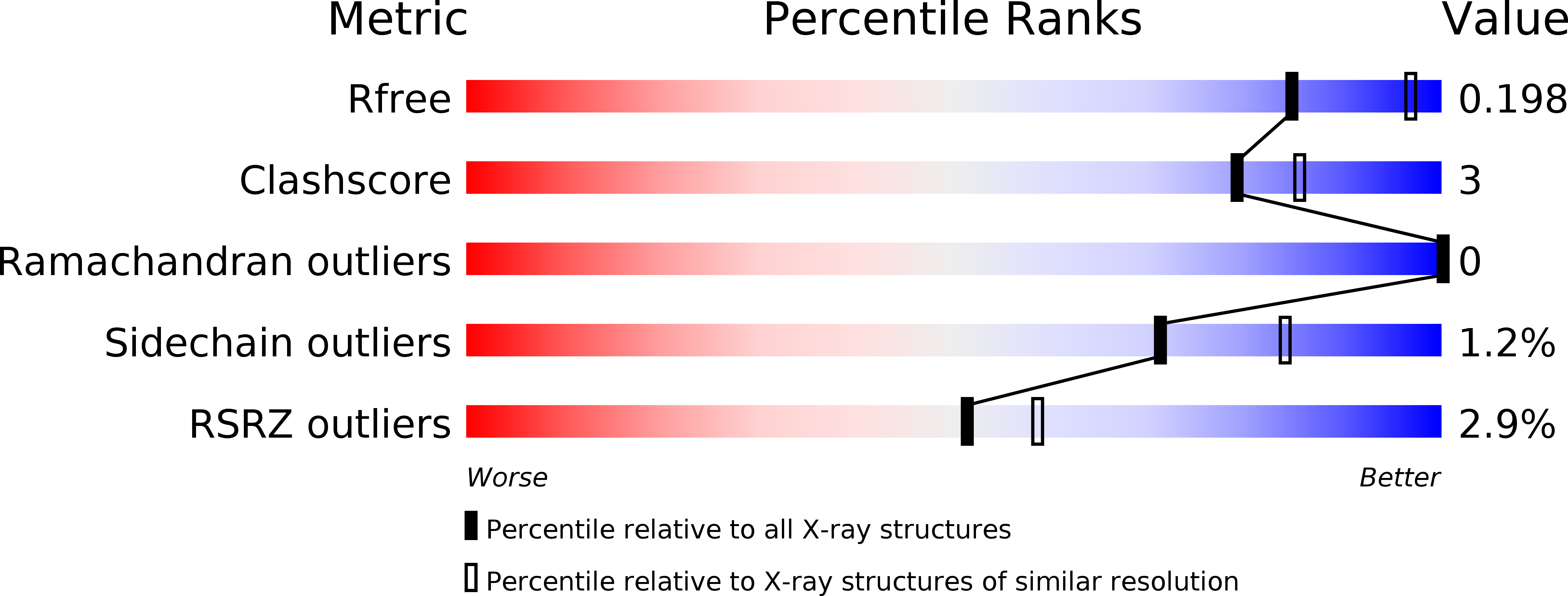
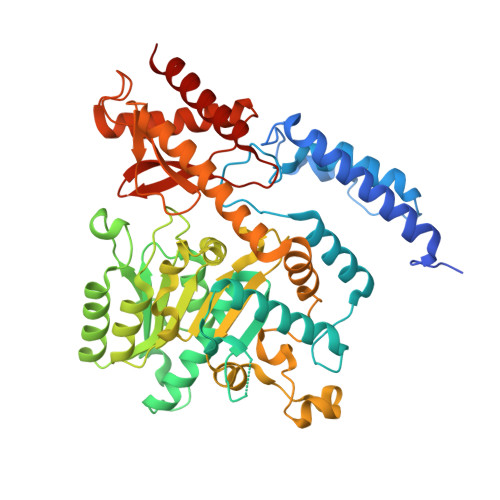
Crystal structure ofOryza sativaTDC reveals the substrate specificity for TDC-mediated melatonin biosynthesis.
Zhou, Y., Liao, L., Liu, X., Liu, B., Chen, X., Guo, Y., Huang, C., Zhao, Y., Zeng, Z.(2020) J Adv Res 24: 501-511
- PubMed: 32595985
- DOI: https://doi.org/10.1016/j.jare.2020.06.004
- Primary Citation of Related Structures:
6KHN, 6KHO, 6KHP - PubMed Abstract:
Plant tryptophan decarboxylase (TDC) is a type II Pyridoxal-5'-phosphate-dependent decarboxylase (PLP_DC) that could be used as a target to genetically improve crops. However, lack of accurate structural information on plant TDC hampers the understanding of its decarboxylation mechanisms. In the present study, the crystal structures of Oryza sativa TDC ( O sTDC) in its complexes with pyridoxal-5'-phosphate, tryptamine and serotonin were determined. The structures provide detailed interaction information between TDC and its substrates. The Y359 residue from the loop gate is a proton donor and forms a Lewis acid-base pair with serotonin/tryptamine, which is associated with product release. The H214 residue is responsible for PLP binding and proton transfer, and its proper interaction with Y359 is essential for Os TDC enzyme activity. The extra hydrogen bonds formed between the 5-hydroxyl group of serotonin and the backbone carboxyl groups of F104 and P105 explain the discrepancy between the catalytic activity of TDC in tryptophan and in 5-hydroxytryptophan. In addition, an evolutionary analysis revealed that type II PLP_DC originated from glutamic acid decarboxylase, potentially as an adaptive evolution of mechanism in organisms in extreme environments. This study is, to our knowledge, the first to present a detailed analysis of the crystal structure of Os TDC in these complexes. The information regarding the catalytic mechanism described here could facilitate the development of protocols to regulate melatonin levels and thereby contribute to crop improvement efforts to improve food security worldwide.
Organizational Affiliation:
National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China.