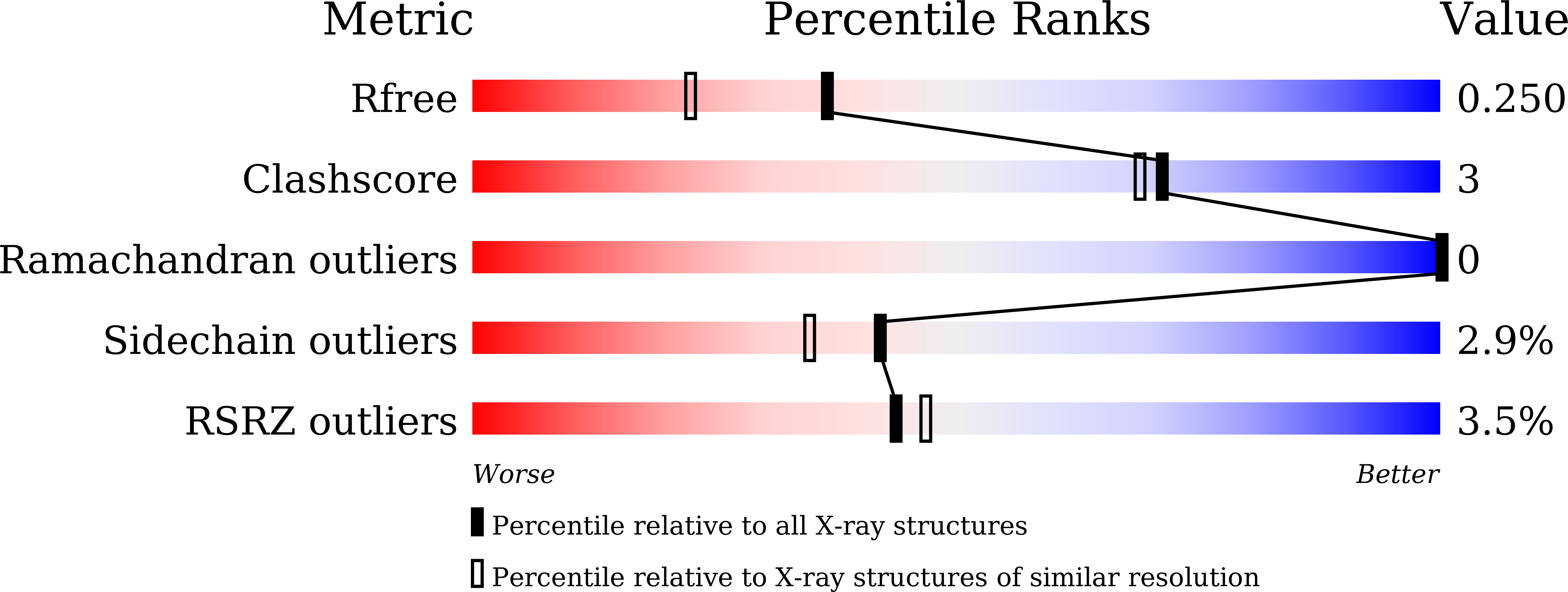
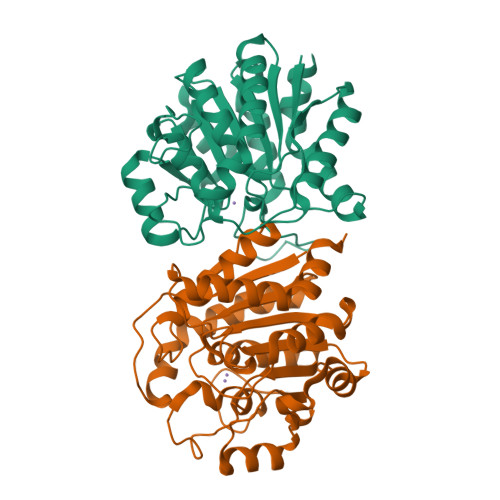
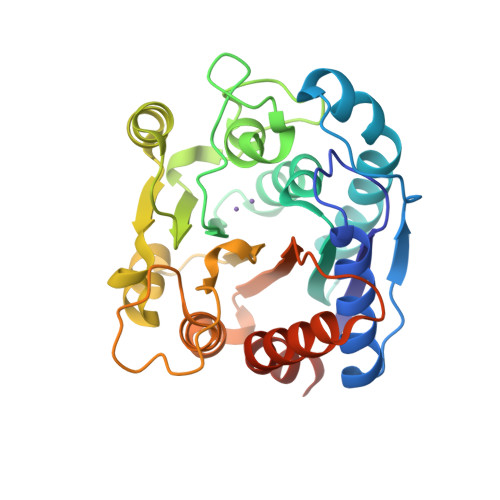
Crystal structure of an Nomega-hydroxy-L-arginine hydrolase found in the D-cycloserine biosynthetic pathway.
Oda, K., Shimotani, N., Kuroda, T., Matoba, Y.(2020) Acta Crystallogr D Struct Biol 76: 506-514
- PubMed: 32496212
- DOI: https://doi.org/10.1107/S2059798320004908
- Primary Citation of Related Structures:
6LUG, 6LUH - PubMed Abstract:
DcsB, one of the enzymes encoded in the D-cycloserine (D-CS) biosynthetic gene cluster, displays a high sequence homology to arginase, which contains two manganese ions in the active site. However, DcsB hydrolyzes N ω -hydroxy-L-arginine, but not L-arginine, to supply hydroxyurea for the biosynthesis of D-CS. Here, the crystal structure of DcsB was determined at a resolution of 1.5 Å using anomalous scattering from the manganese ions. In the crystal structure, DscB generates an artificial dimer created by the open and closed forms. Gel-filtration analysis demonstrated that DcsB is a monomeric protein, unlike arginase, which forms a trimeric structure. The active center containing the binuclear manganese cluster differs between DcsB and arginase. In DcsB, one of the ligands of the Mn A ion is a cysteine, while the corresponding residue in arginase is a histidine. In addition, DcsB has no counterpart to the histidine residue that acts as a general acid/base during the catalytic reaction of arginase. The present study demonstrates that DcsB has a unique active site that differs from that of arginase.
Organizational Affiliation:
Department of Virology, Institute of Biomedical and Health Sciences, Hiroshima University, 1-2-3 Kasumi, Minami-ku, Hiroshima 734-8553, Japan.