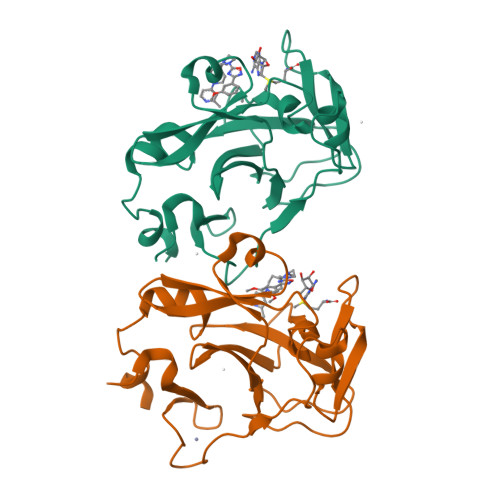
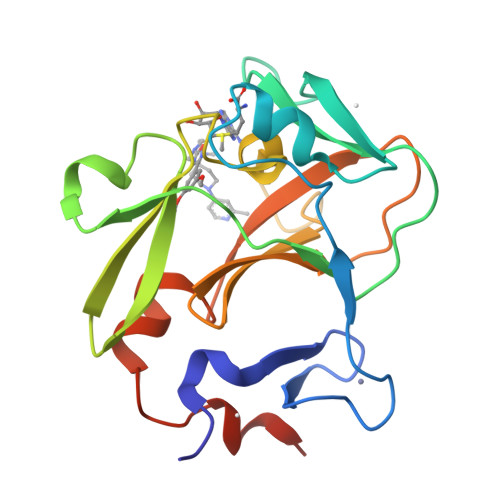
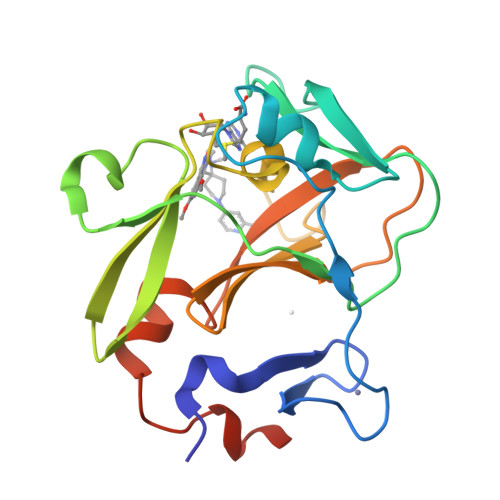
Discovery of a chemical probe for PRDM9.
Allali-Hassani, A., Szewczyk, M.M., Ivanochko, D., Organ, S.L., Bok, J., Ho, J.S.Y., Gay, F.P.H., Li, F., Blazer, L., Eram, M.S., Halabelian, L., Dilworth, D., Luciani, G.M., Lima-Fernandes, E., Wu, Q., Loppnau, P., Palmer, N., Talib, S.Z.A., Brown, P.J., Schapira, M., Kaldis, P., O'Hagan, R.C., Guccione, E., Barsyte-Lovejoy, D., Arrowsmith, C.H., Sanders, J.M., Kattar, S.D., Bennett, D.J., Nicholson, B., Vedadi, M.(2019) Nat Commun 10: 5759-5759
- PubMed: 31848333
- DOI: https://doi.org/10.1038/s41467-019-13652-x
- Primary Citation of Related Structures:
6NM4 - PubMed Abstract:
PRDM9 is a PR domain containing protein which trimethylates histone 3 on lysine 4 and 36. Its normal expression is restricted to germ cells and attenuation of its activity results in altered meiotic gene transcription, impairment of double-stranded breaks and pairing between homologous chromosomes. There is growing evidence for a role of aberrant expression of PRDM9 in oncogenesis and genome instability. Here we report the discovery of MRK-740, a potent (IC 50 : 80 ± 16 nM), selective and cell-active PRDM9 inhibitor (Chemical Probe). MRK-740 binds in the substrate-binding pocket, with unusually extensive interactions with the cofactor S-adenosylmethionine (SAM), conferring SAM-dependent substrate-competitive inhibition. In cells, MRK-740 specifically and directly inhibits H3K4 methylation at endogenous PRDM9 target loci, whereas the closely related inactive control compound, MRK-740-NC, does not. The discovery of MRK-740 as a chemical probe for the PRDM subfamily of methyltransferases highlights the potential for exploiting SAM in targeting SAM-dependent methyltransferases.
Organizational Affiliation:
Structural Genomics Consortium, University of Toronto, Toronto, ON, M5G 1L7, Canada.