Understanding the structural basis of species selective, stereospecific inhibition for Cryptosporidium and human thymidylate synthase.
Czyzyk, D.J., Valhondo, M., Jorgensen, W.L., Anderson, K.S.(2019) FEBS Lett 593: 2069-2078
- PubMed: 31172516
- DOI: https://doi.org/10.1002/1873-3468.13474
- Primary Citation of Related Structures:
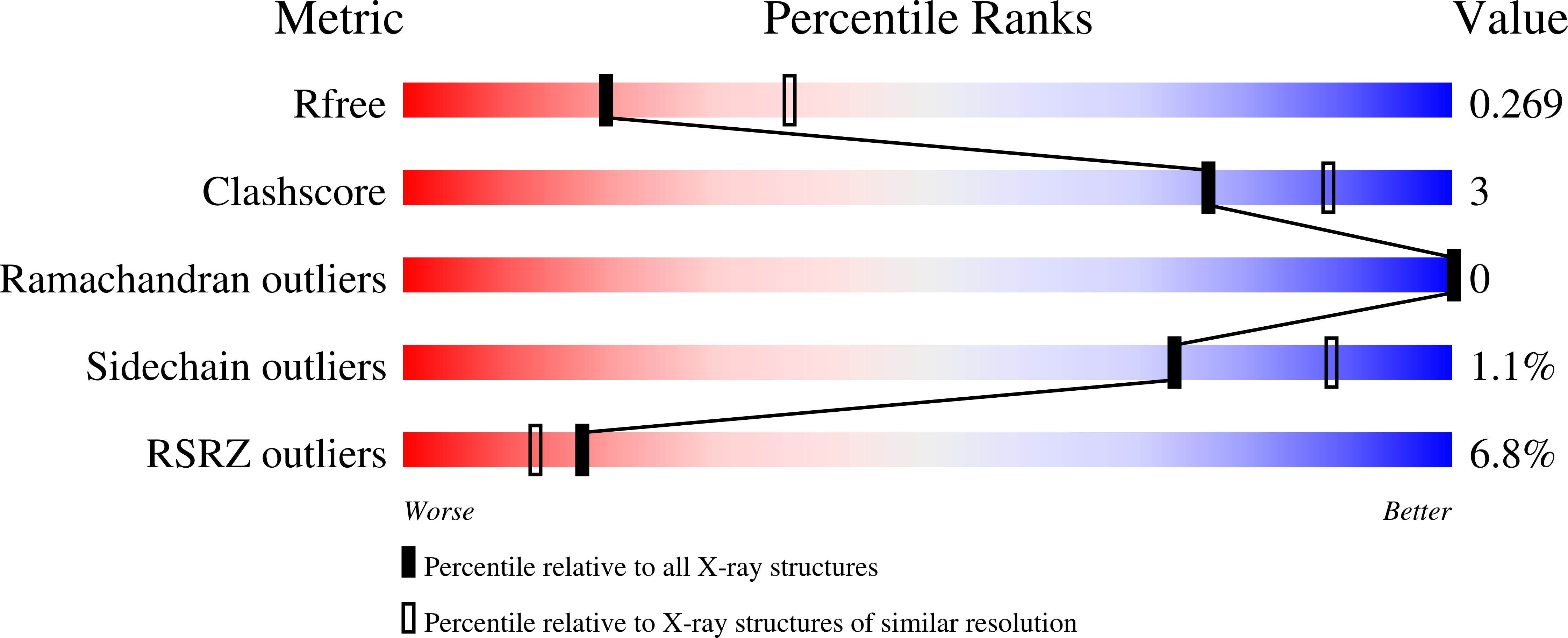
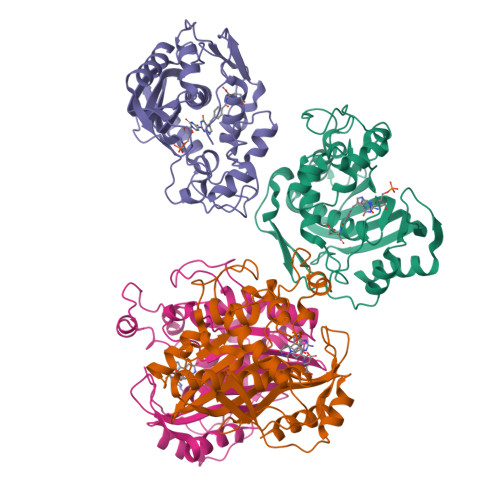
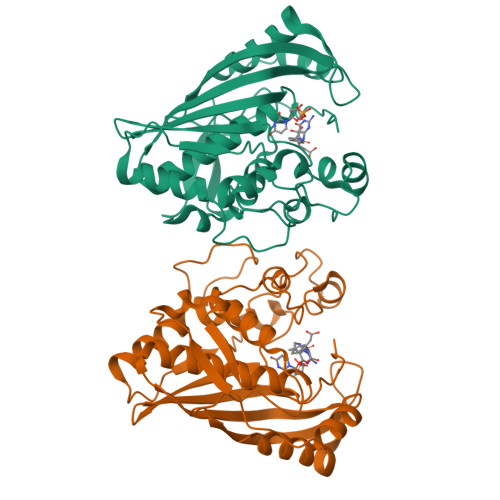
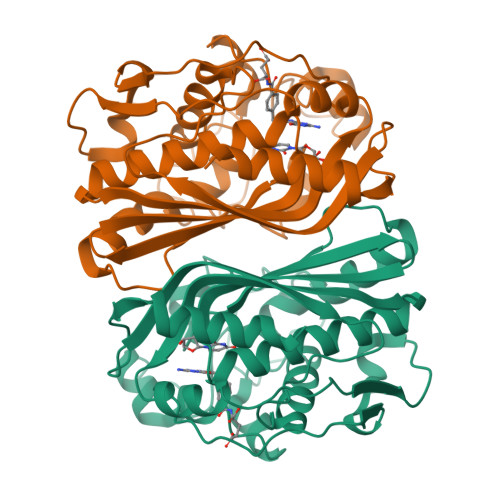
6OJS, 6OJU, 6OJV - PubMed Abstract:
Thymidylate synthase (TS), found in all organisms, is an essential enzyme responsible for the de novo synthesis of deoxythymidine monophosphate. The TS active sites of the protozoal parasite Cryptosporidium hominis and human are relatively conserved. Evaluation of antifolate compound 1 and its R-enantiomer 2 against both enzymes reveals divergent inhibitor selectivity and enzyme stereospecificity. To establish how C. hominis and human TS (ChTS and hTS) selectively discriminate 1 and 2, respectively, we determined crystal structures of ChTS complexed with 2 and hTS complexed with 1 or 2. Coupled with the previously determined structure of ChTS complexed with 1, we discuss a possible mechanism for enzyme stereospecificity and inhibitor selectivity.
Organizational Affiliation:
Department of Pharmacology, Yale University School of Medicine, New Haven, CT, USA.