Conservation of Atypical Allostery inC. elegansUDP-Glucose Dehydrogenase.
Beattie, N.R., Keul, N.D., Hicks Sirmans, T.N., McDonald, W.E., Talmadge, T.M., Taujale, R., Kannan, N., Wood, Z.A.(2019) ACS Omega 4: 16318-16329
- PubMed: 31616809
- DOI: https://doi.org/10.1021/acsomega.9b01565
- Primary Citation of Related Structures:
6OM8 - PubMed Abstract:
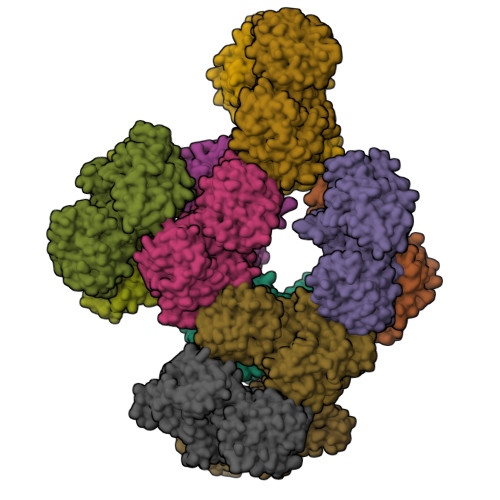
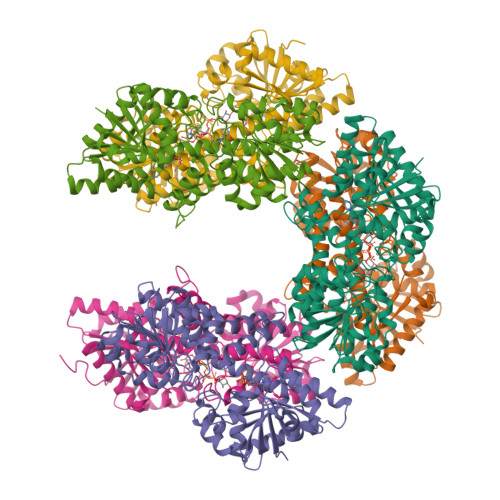
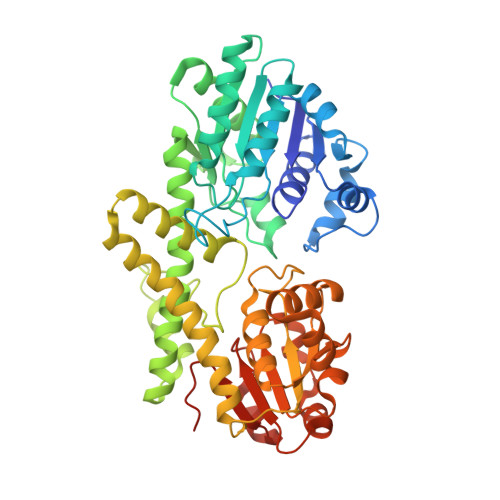
Human UDP-glucose dehydrogenase (hUGDH) oxidizes uridine diphosphate (UDP)-glucose to UDP-glucuronic acid, an essential substrate in the phase II metabolism of drugs. The activity of hUGDH is controlled by an atypical allosteric mechanism in which the feedback inhibitor UDP-xylose competes with the substrate for the active site and triggers a buried allosteric switch to produce an inactive complex (E Ω ). Previous comparisons with a nonallosteric UGDH identified six large-to-small substitutions that produce packing defects in the protein core and provide the conformational flexibility necessary for the allosteric transition. Here, we test the hypothesis that these large-to-small substitutions form a motif that can be used to identify allosteric UGDHs. Caenorhabditis elegans UGDH (cUGDH) conserves this motif with the exception of an Ala-to-Pro substitution in position 109. The crystal structures of unliganded and UDP-xylose bound cUGDH show that the A109P substitution is accommodated by an Asn-to-Ser substitution at position 290. Steady-state analysis and sedimentation velocity studies show that the allosteric transition is conserved in cUGDH. The enzyme also exhibits hysteresis in progress curves and negative cooperativity with respect to NAD + binding. Both of these phenomena are conserved in the human enzyme, which is strong evidence that these represent fundamental features of atypical allostery in UGDH. A phylogenetic analysis of UGDH shows that the atypical allostery motif is ancient and identifies a potential transition point in the evolution of the UGDH family.
Organizational Affiliation:
Department of Biochemistry & Molecular Biology and Institute of Bioinformatics, University of Georgia, Athens, Georgia 30602, United States.