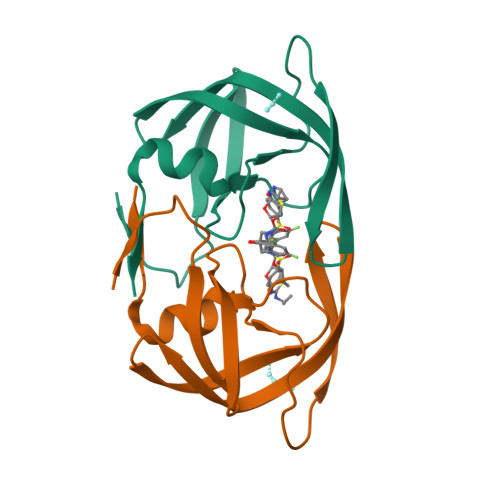
Potent antiviral HIV-1 protease inhibitor combats highly drug resistant mutant PR20.
Kneller, D.W., Agniswamy, J., Ghosh, A.K., Weber, I.T.(2019) Biochem Biophys Res Commun 519: 61-66
- PubMed: 31474336
- DOI: https://doi.org/10.1016/j.bbrc.2019.08.126
- Primary Citation of Related Structures:
6PRF - PubMed Abstract:
Drug-resistance threatens effective treatment of HIV/AIDS. Clinical inhibitors, including darunavir (1), are ineffective for highly resistant protease mutant PR20, however, antiviral compound 2 derived from 1 with fused tricyclic group at P2, extended amino-benzothiazole P2' ligand and two fluorine atoms on P1 shows 16-fold better inhibition of PR20 enzyme activity. Crystal structures of PR20 and wild-type PR complexes reveal how the extra groups of 2 counteract the expanded ligand-binding pocket, dynamic flaps, and faster dimer dissociation of PR20.
Organizational Affiliation:
Department of Biology, Georgia State University, Atlanta, GA, 30303, USA.