Discovery ofN-Substituted 3-Amino-4-(3-boronopropyl)pyrrolidine-3-carboxylic Acids as Highly Potent Third-Generation Inhibitors of Human Arginase I and II.
Van Zandt, M.C., Jagdmann, G.E., Whitehouse, D.L., Ji, M., Savoy, J., Potapova, O., Cousido-Siah, A., Mitschler, A., Howard, E.I., Pyle, A.M., Podjarny, A.D.(2019) J Med Chem 62: 8164-8177
- PubMed: 31408339
- DOI: https://doi.org/10.1021/acs.jmedchem.9b00931
- Primary Citation of Related Structures:
6Q37, 6Q39 - PubMed Abstract:
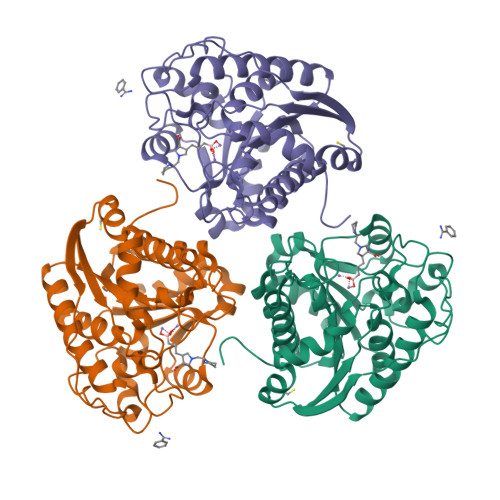
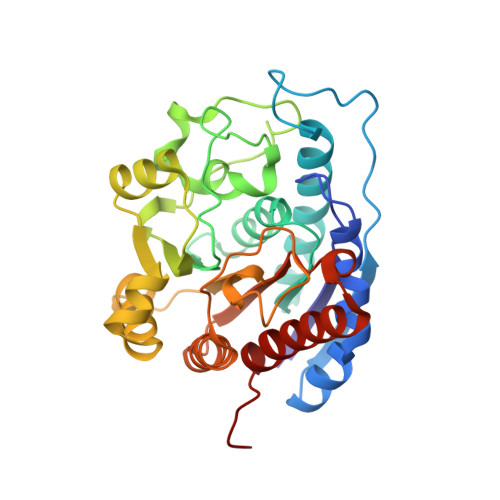
Recent efforts to identify new highly potent arginase inhibitors have resulted in the discovery of a novel family of (3 R ,4 S )-3-amino-4-(3-boronopropyl)pyrrolidine-3-carboxylic acid analogues with up to a 1000-fold increase in potency relative to the current standards, 2-amino-6-boronohexanoic acid (ABH) and N -hydroxy-nor-l-arginine (nor-NOHA). The lead candidate, with an N -2-amino-3-phenylpropyl substituent (NED-3238), example 43 , inhibits arginase I and II with IC 50 values of 1.3 and 8.1 nM, respectively. Herein, we report the design, synthesis, and structure-activity relationships for this novel series of inhibitors, along with X-ray crystallographic data for selected examples bound to human arginase II.
Organizational Affiliation:
New England Discovery Partners , 23 Business Park Drive , Branford , Connecticut 06405 , United States.