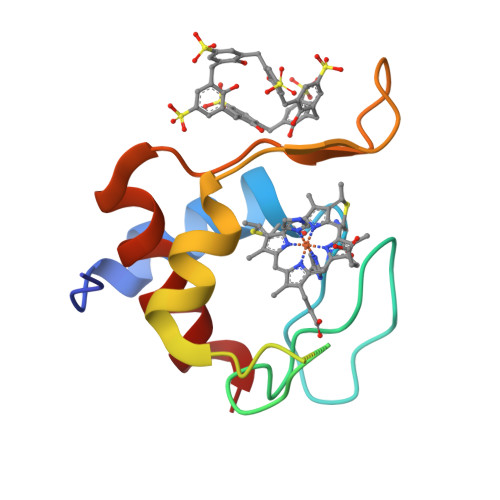
Calixarene capture of partially unfolded cytochrome c.
Engilberge, S., Rennie, M.L., Crowley, P.B.(2019) FEBS Lett 593: 2112-2117
- PubMed: 31254353
- DOI: https://doi.org/10.1002/1873-3468.13512
- Primary Citation of Related Structures:
6RGI - PubMed Abstract:
Supramolecular receptors such as water-soluble calixarenes are in development as 'molecular glues' for protein assembly. Here, we obtained cocrystals of sulfonato-calix[6]arene (sclx 6 ) and yeast cytochrome c (cytc) in the presence of imidazole. A crystal structure at 2.65 Å resolution reveals major structural rearrangement and disorder in imidazole-bound cytc. The largest protein-calixarene interface involves 440 Å 2 of the protein surface with key contacts at Arg13, Lys73, and Lys79. These lysines participate in alkaline transitions of cytc and are part of Ω-loop D, which is substantially restructured in the complex with sclx 6 . The structural modification also includes Ω-loop C, which is disordered (residues 41-55 inclusive). These results suggest the possibility of using supramolecular scaffolds to trap partially disordered proteins.
Organizational Affiliation:
School of Chemistry, National University of Ireland Galway, Galway, Ireland.