Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
Armstrong, Z., Kuo, C.L., Lahav, D., Liu, B., Johnson, R., Beenakker, T.J.M., de Boer, C., Wong, C.S., van Rijssel, E.R., Debets, M.F., Florea, B.I., Hissink, C., Boot, R.G., Geurink, P.P., Ovaa, H., van der Stelt, M., van der Marel, G.M., Codee, J.D.C., Aerts, J.M.F.G., Wu, L., Overkleeft, H.S., Davies, G.J.(2020) J Am Chem Soc 142: 13021-13029
- PubMed: 32605368
- DOI: https://doi.org/10.1021/jacs.0c03880
- Primary Citation of Related Structures:
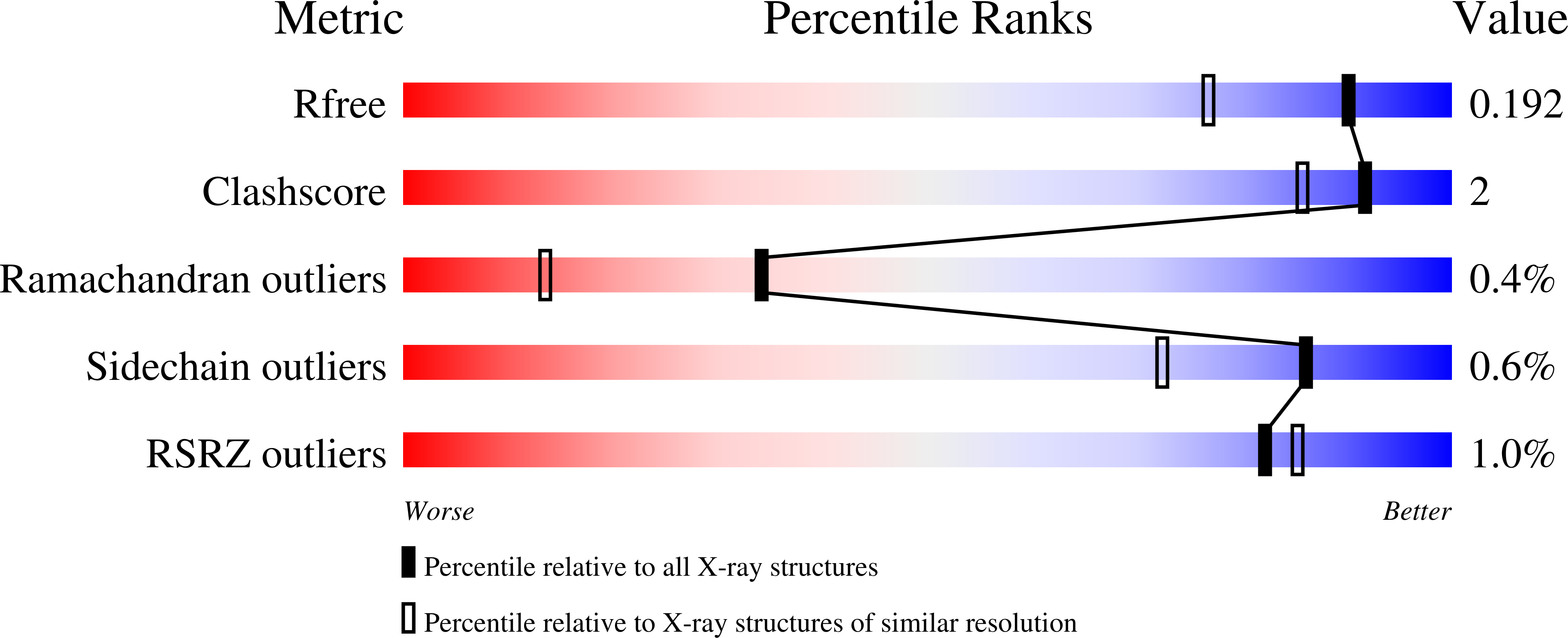
6RPC, 6RQZ, 6RRH, 6RRJ, 6RRN, 6RRU, 6RRW, 6RRX, 6RRY, 6RS0 - PubMed Abstract:
Golgi mannosidase II (GMII) catalyzes the sequential hydrolysis of two mannosyl residues from GlcNAcMan 5 GlcNAc 2 to produce GlcNAcMan 3 GlcNAc 2 , the precursor for all complex N -glycans, including the branched N -glycans associated with cancer. Inhibitors of GMII are potential cancer therapeutics, but their usefulness is limited by off-target effects, which produce α-mannosidosis-like symptoms. Despite many structural and mechanistic studies of GMII, we still lack a potent and selective inhibitor of this enzyme. Here, we synthesized manno- epi -cyclophellitol epoxide and aziridines and demonstrate their covalent modification and time-dependent inhibition of GMII. Application of fluorescent manno- epi -cyclophellitol aziridine derivatives enabled activity-based protein profiling of α-mannosidases from both human cell lysate and mouse tissue extracts. Synthesized probes also facilitated a fluorescence polarization-based screen for dGMII inhibitors. We identified seven previously unknown inhibitors of GMII from a library of over 350 iminosugars and investigated their binding modalities through X-ray crystallography. Our results reveal previously unobserved inhibitor binding modes and promising scaffolds for the generation of selective GMII inhibitors.
Organizational Affiliation:
Structural Biology Laboratory, Department of Chemistry, The University of York, York YO10 5DD, United Kingdom.