Profiling Substrate Promiscuity of Wild-Type Sugar Kinases for Multi-fluorinated Monosaccharides.
Keenan, T., Parmeggiani, F., Malassis, J., Fontenelle, C.Q., Vendeville, J.B., Offen, W., Both, P., Huang, K., Marchesi, A., Heyam, A., Young, C., Charnock, S.J., Davies, G.J., Linclau, B., Flitsch, S.L., Fascione, M.A.(2020) Cell Chem Biol 27: 1199
- PubMed: 32619452
- DOI: https://doi.org/10.1016/j.chembiol.2020.06.005
- Primary Citation of Related Structures:
6TEP, 6TEQ, 6TER - PubMed Abstract:
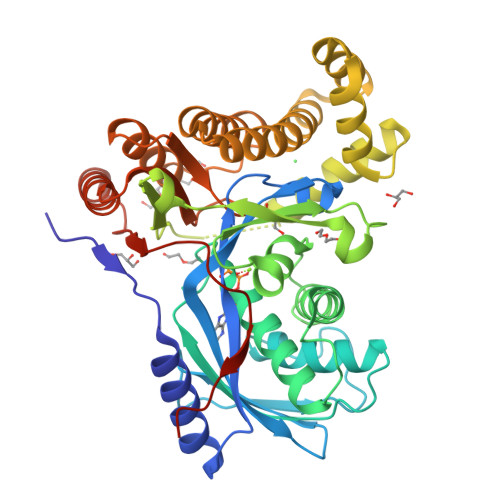
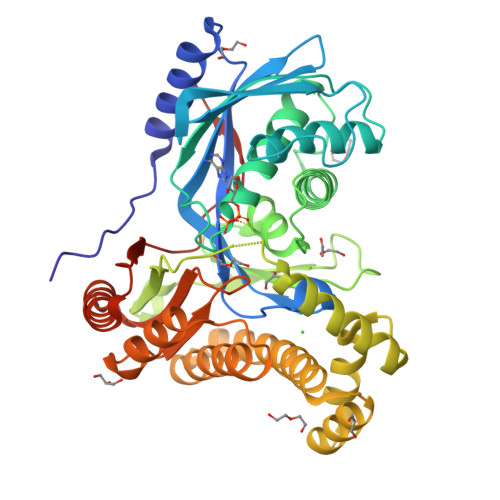
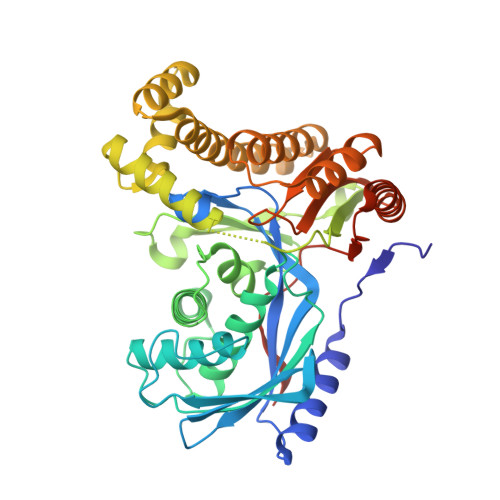
Fluorinated sugar-1-phosphates are of emerging importance as intermediates in the chemical and biocatalytic synthesis of modified oligosaccharides, as well as probes for chemical biology. Here we present a systematic study of the activity of a wide range of anomeric sugar kinases (galacto- and N-acetylhexosamine kinases) against a panel of fluorinated monosaccharides, leading to the first examples of polyfluorinated substrates accepted by this class of enzymes. We have discovered four new N-acetylhexosamine kinases with a different substrate scope, thus expanding the number of homologs available in this subclass of kinases. Lastly, we have solved the crystal structure of a galactokinase in complex with 2-deoxy-2-fluorogalactose, giving insight into changes in the active site that may account for the specificity of the enzyme toward certain substrate analogs.
Organizational Affiliation:
Department of Chemistry, University of York, Heslington, York YO10 5DD, UK.