Dismantling and Rebuilding the Trisulfide Cofactor Demonstrates Its Essential Role in Human Sulfide Quinone Oxidoreductase.
Landry, A.P., Moon, S., Bonanata, J., Cho, U.S., Coitino, E.L., Banerjee, R.(2020) J Am Chem Soc 142: 14295-14306
- PubMed: 32787249
- DOI: https://doi.org/10.1021/jacs.0c06066
- Primary Citation of Related Structures:
6WH6 - PubMed Abstract:
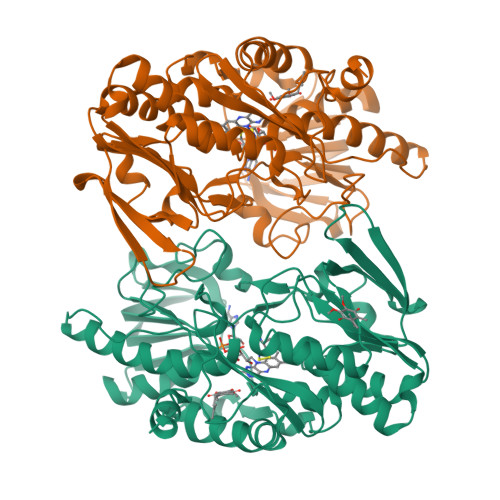
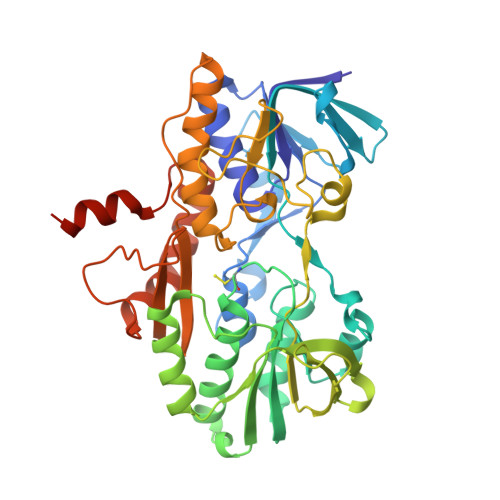
Sulfide quinone oxidoreductase (SQOR) catalyzes the first step in sulfide clearance, coupling H 2 S oxidation to coenzyme Q reduction. Recent structures of human SQOR revealed a sulfur atom bridging the SQOR active site cysteines in a trisulfide configuration. Here, we assessed the importance of this cofactor using kinetic, crystallographic, and computational modeling approaches. Cyanolysis of SQOR proceeds via formation of an intense charge transfer complex that subsequently decays to eliminate thiocyanate. We captured a disulfanyl-methanimido thioate intermediate in the SQOR crystal structure, revealing how cyanolysis leads to reversible loss of SQOR activity that is restored in the presence of sulfide. Computational modeling and MD simulations revealed an ∼10 5 -fold rate enhancement for nucleophilic addition of sulfide into the trisulfide versus a disulfide cofactor. The cysteine trisulfide in SQOR is thus critical for activity and provides a significant catalytic advantage over a cysteine disulfide.
Organizational Affiliation:
Department of Biological Chemistry, University of Michigan Medical School, Ann Arbor, Michigan 48109, United States.