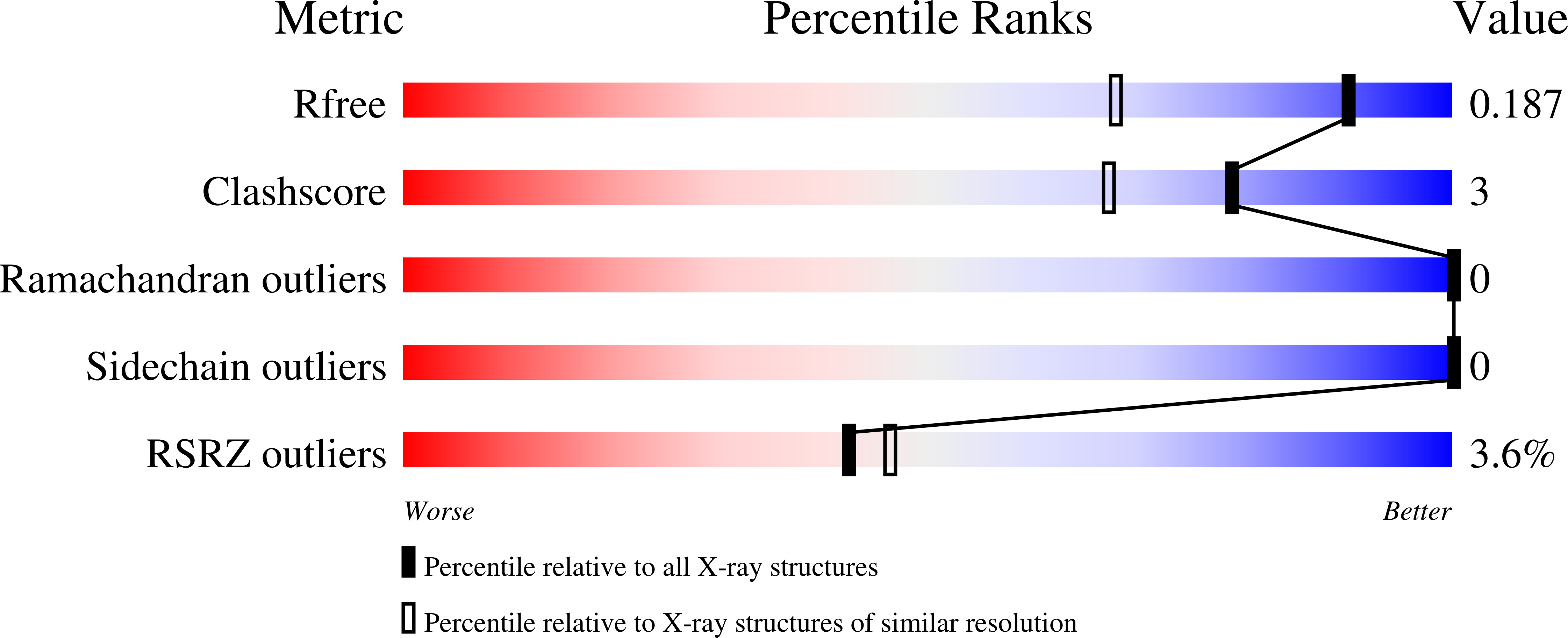
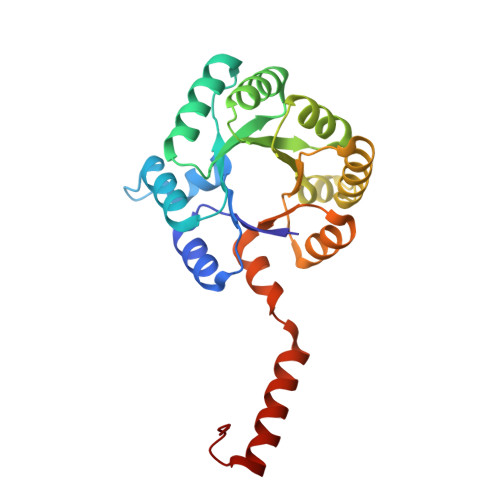
Large-scale motions underlie physical but not chemical steps in transaldolase mechanism: Substrate binding by conformational selection and rate-determining product release
Sautner, V., Lietzow, T.H., Klaus, M., Funk, L.M., Tittmann, K.To be published.