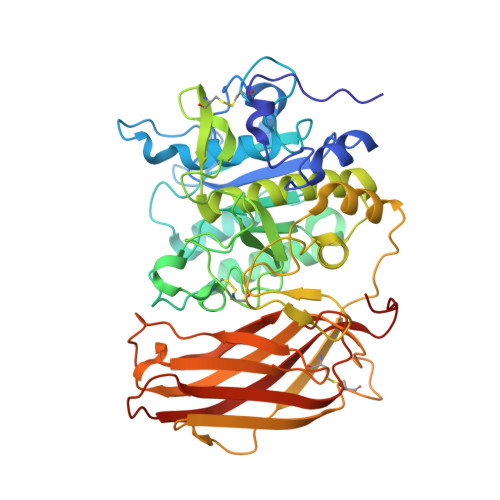
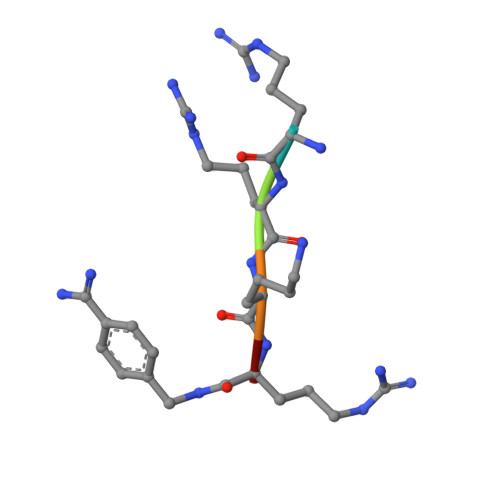
Design, Synthesis, and Characterization of Macrocyclic Inhibitors of the Proprotein Convertase Furin.
Van Lam van, T., Ivanova, T., Hardes, K., Heindl, M.R., Morty, R.E., Bottcher-Friebertshauser, E., Lindberg, I., Than, M.E., Dahms, S.O., Steinmetzer, T.(2019) ChemMedChem 14: 673-685
- PubMed: 30680958
- DOI: https://doi.org/10.1002/cmdc.201800807
- Primary Citation of Related Structures:
6HLB, 6HLD, 6HLE, 6HZA, 6HZB, 6HZC, 6HZD - PubMed Abstract:
The activation of viral glycoproteins by the host protease furin is an essential step in the replication of numerous pathogenic viruses. Thus, effective inhibitors of furin could serve as broad-spectrum antiviral drugs. A crystal structure of an inhibitory hexapeptide derivative in complex with furin served as template for the rational design of various types of new cyclic inhibitors. Most of the prepared derivatives are relatively potent furin inhibitors with inhibition constants in the low nanomolar or even sub-nanomolar range. For seven derivatives the crystal structures in complex with furin could be determined. In three complexes, electron density was found for the entire inhibitor. In the other cases the structures could be determined only for the P6/P5-P1 segments, which directly interact with furin. The cyclic derivatives together with two non-cyclic reference compounds were tested as inhibitors of the proteolytic activation and replication of respiratory syncytial virus in cells. Significant antiviral activity was found for both linear reference inhibitors, whereas a negligible efficacy was determined for the cyclic derivatives.
Organizational Affiliation:
Institute of Pharmaceutical Chemistry, Philipps University, Marbacher Weg 6, 35032, Marburg, Germany.