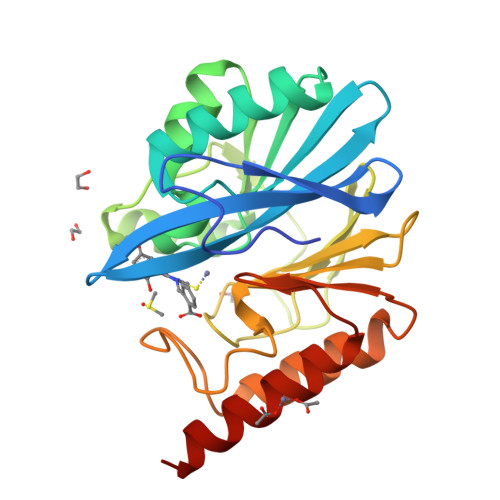
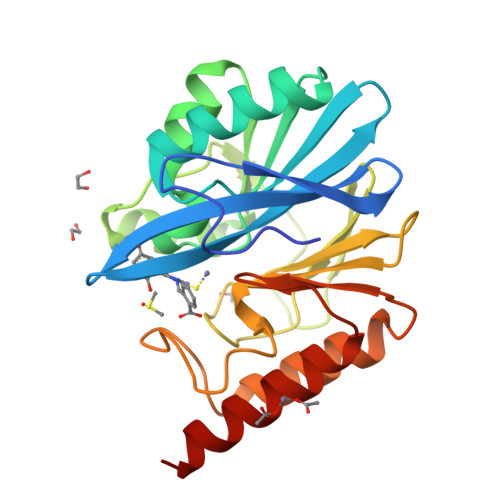
4-Amino-1,2,4-triazole-3-thione-derived Schiff bases as metallo-beta-lactamase inhibitors.
Gavara, L., Sevaille, L., De Luca, F., Mercuri, P., Bebrone, C., Feller, G., Legru, A., Cerboni, G., Tanfoni, S., Baud, D., Cutolo, G., Bestgen, B., Chelini, G., Verdirosa, F., Sannio, F., Pozzi, C., Benvenuti, M., Kwapien, K., Fischer, M., Becker, K., Frere, J.M., Mangani, S., Gresh, N., Berthomieu, D., Galleni, M., Docquier, J.D., Hernandez, J.F.(2020) Eur J Med Chem 208: 112720-112720
- PubMed: 32937203
- DOI: https://doi.org/10.1016/j.ejmech.2020.112720
- Primary Citation of Related Structures:
6YRP - PubMed Abstract:
Resistance to β-lactam antibiotics in Gram-negatives producing metallo-β-lactamases (MBLs) represents a major medical threat and there is an extremely urgent need to develop clinically useful inhibitors. We previously reported the original binding mode of 5-substituted-4-amino/H-1,2,4-triazole-3-thione compounds in the catalytic site of an MBL. Moreover, we showed that, although moderately potent, they represented a promising basis for the development of broad-spectrum MBL inhibitors. Here, we synthesized and characterized a large number of 4-amino-1,2,4-triazole-3-thione-derived Schiff bases. Compared to the previous series, the presence of an aryl moiety at position 4 afforded an average 10-fold increase in potency. Among 90 synthetic compounds, more than half inhibited at least one of the six tested MBLs (L1, VIM-4, VIM-2, NDM-1, IMP-1, CphA) with K i values in the μM to sub-μM range. Several were broad-spectrum inhibitors, also inhibiting the most clinically relevant VIM-2 and NDM-1. Active compounds generally contained halogenated, bicyclic aryl or phenolic moieties at position 5, and one substituent among o-benzoic, 2,4-dihydroxyphenyl, p-benzyloxyphenyl or 3-(m-benzoyl)-phenyl at position 4. The crystallographic structure of VIM-2 in complex with an inhibitor showed the expected binding between the triazole-thione moiety and the dinuclear centre and also revealed a network of interactions involving Phe61, Tyr67, Trp87 and the conserved Asn233. Microbiological analysis suggested that the potentiation activity of the compounds was limited by poor outer membrane penetration or efflux. This was supported by the ability of one compound to restore the susceptibility of an NDM-1-producing E. coli clinical strain toward several β-lactams in the presence only of a sub-inhibitory concentration of colistin, a permeabilizing agent. Finally, some compounds were tested against the structurally similar di-zinc human glyoxalase II and found weaker inhibitors of the latter enzyme, thus showing a promising selectivity towards MBLs.
Organizational Affiliation:
Institut des Biomolécules Max Mousseron, UMR5247 CNRS, Université de Montpellier, ENSCM, Faculté de Pharmacie, 34093, Montpellier Cedex 5, France. Electronic address: laurent.gavara@umontpellier.fr.