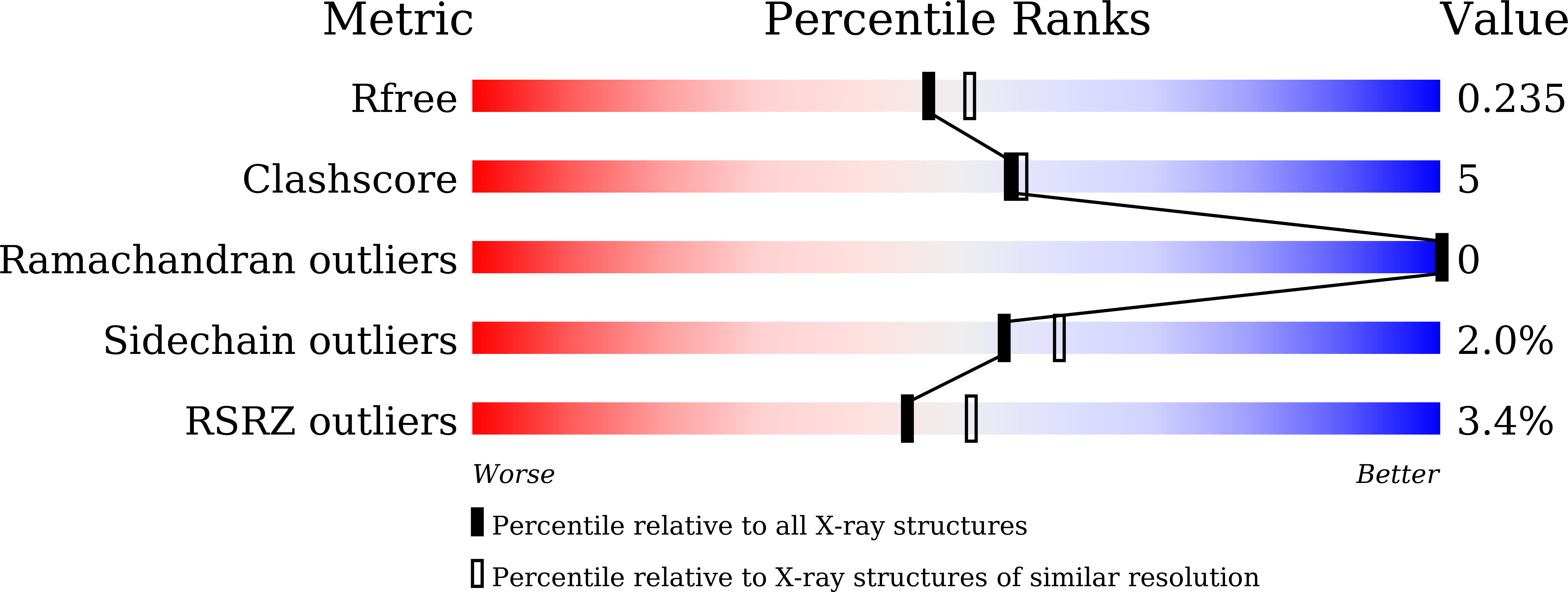
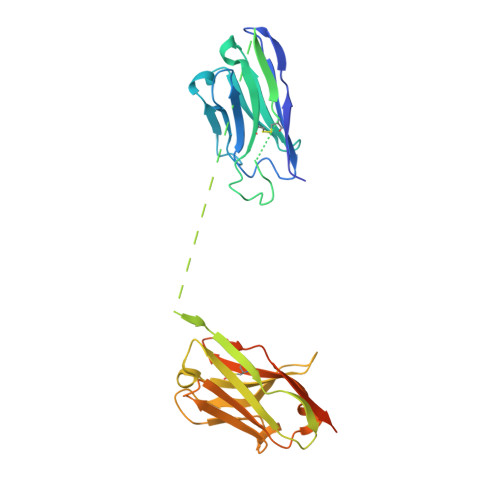
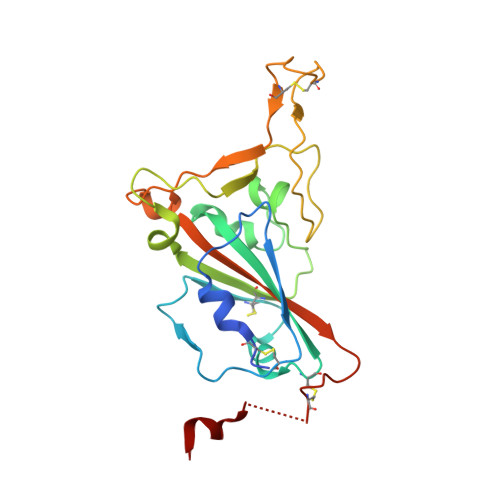
A high-affinity RBD-targeting nanobody improves fusion partner's potency against SARS-CoV-2.
Yao, H., Cai, H., Li, T., Zhou, B., Qin, W., Lavillette, D., Li, D.(2021) PLoS Pathog 17: e1009328-e1009328
- PubMed: 33657135
- DOI: https://doi.org/10.1371/journal.ppat.1009328
- Primary Citation of Related Structures:
7D2Z, 7D30 - PubMed Abstract:
A key step to the SARS-CoV-2 infection is the attachment of its Spike receptor-binding domain (S RBD) to the host receptor ACE2. Considerable research has been devoted to the development of neutralizing antibodies, including llama-derived single-chain nanobodies, to target the receptor-binding motif (RBM) and to block ACE2-RBD binding. Simple and effective strategies to increase potency are desirable for such studies when antibodies are only modestly effective. Here, we identify and characterize a high-affinity synthetic nanobody (sybody, SR31) as a fusion partner to improve the potency of RBM-antibodies. Crystallographic studies reveal that SR31 binds to RBD at a conserved and 'greasy' site distal to RBM. Although SR31 distorts RBD at the interface, it does not perturb the RBM conformation, hence displaying no neutralizing activities itself. However, fusing SR31 to two modestly neutralizing sybodies dramatically increases their affinity for RBD and neutralization activity against SARS-CoV-2 pseudovirus. Our work presents a tool protein and an efficient strategy to improve nanobody potency.
Organizational Affiliation:
CAS Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, Shanghai, China.