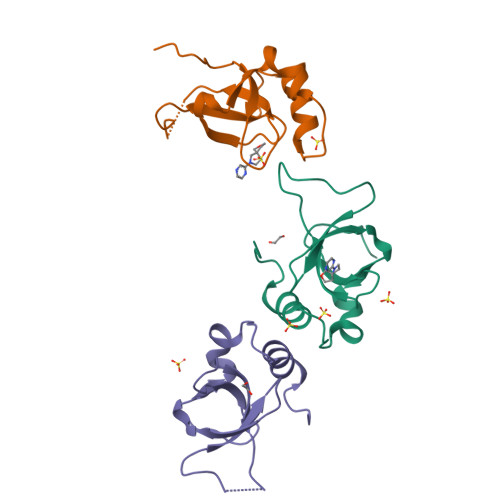
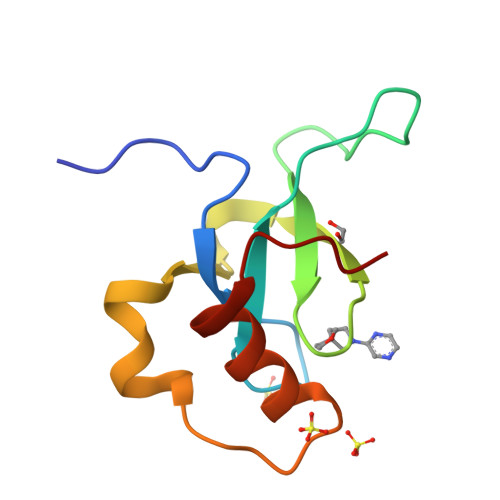
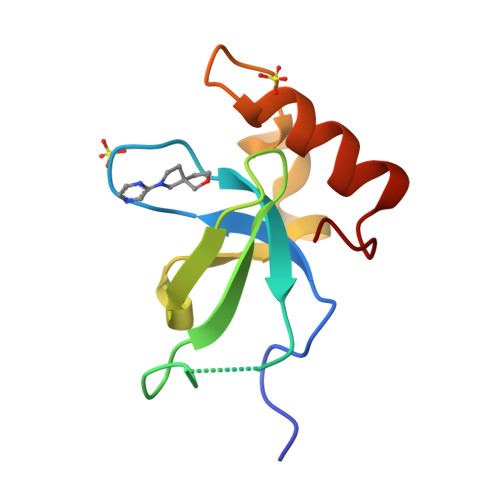
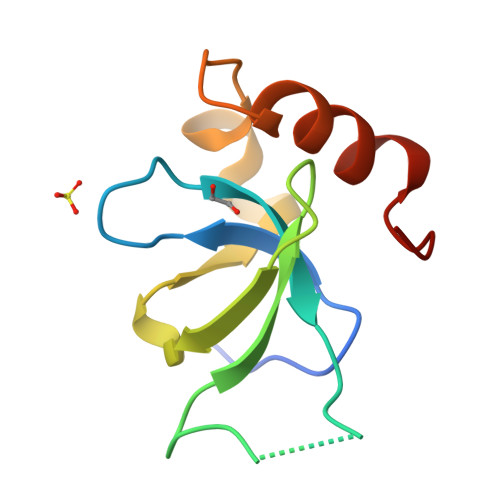
Rational fragment-based design of compounds targeting the PWWP domain of the HRP family.
Vantieghem, T., Aslam, N.A., Osipov, E.M., Akele, M., Van Belle, S., Beelen, S., Drexler, M., Paulovcakova, T., Lux, V., Fearon, D., Douangamath, A., von Delft, F., Christ, F., Veverka, V., Verwilst, P., Van Aerschot, A., Debyser, Z., Strelkov, S.V.(2024) Eur J Med Chem 280: 116960-116960
- PubMed: 39461037
- DOI: https://doi.org/10.1016/j.ejmech.2024.116960
- Primary Citation of Related Structures:
7HG0, 7HG1, 7HG2, 7HG3, 7HG4, 7HG5, 7HG6, 7HG7, 7HG8, 7HG9, 7HGA, 7HGB, 7HGC, 7HGD, 7HGE, 7HGF, 7HGG, 7HGH, 7HGI, 7HGJ, 7HGK, 7HGL, 7HGM, 7HGN, 7HGO, 7HGP, 7HGQ, 7HGR, 7HGS, 7HGT, 7HGU, 7HGV, 7HGW, 7HGX, 7HGY, 7HGZ, 7HH0, 7HH1, 7HH2, 7HH3, 7HH4, 7HH5, 7HH6, 7HH7, 7HH8, 7HH9, 7HHA, 7HHB, 7HHC, 7HHD - PubMed Abstract:
Lens epithelium-derived growth factor p75 (LEDGF/p75), member of the hepatoma-derived growth-factor-related protein (HRP) family, is a transcriptional co-activator and involved in several pathologies including HIV infection and malignancies such as MLL-rearranged leukemia. LEDGF/p75 acts by tethering proteins to the chromatin through its integrase binding domain. This chromatin interaction occurs between the PWWP domain of LEDGF/p75 and nucleosomes carrying a di- or trimethylation mark on histone H3 Lys36 (H3K36me2/3). Our aim is to rationally devise small molecule drugs capable of inhibiting such interaction. To bootstrap this development, we resorted to X-ray crystallography-based fragment screening (FBS-X). Given that the LEDGF PWWP domain crystals were not suitable for FBS-X, we employed crystals of the closely related PWWP domain of paralog HRP-2. As a result, as many as 68 diverse fragment hits were identified, providing a detailed sampling of the H3K36me2/3 pocket pharmacophore. Subsequent structure-guided fragment expansion in three directions yielded multiple compound series binding to the pocket, as verified through X-ray crystallography, nuclear magnetic resonance and differential scanning fluorimetry. Our best compounds have double-digit micromolar affinity and optimally sample the interactions available in the pocket, judging by the K d -based ligand efficiency exceeding 0.5 kcal/mol per non-hydrogen atom. Beyond π-stacking within the aromatic cage of the pocket and hydrogen bonding, the best compounds engage in a σ-hole interaction between a halogen atom and a conserved water buried deep in the pocket. Notably, the binding pocket in LEDGF PWWP is considerably smaller compared to the related PWWP1 domains of NSD2 and NSD3 which feature an additional subpocket and for which nanomolar affinity compounds have been developed recently. The absence of this subpocket in LEDGF PWWP limits the attainable affinity. Additionally, these structural differences in the H3K36me2/3 pocket across the PWWP domain family translate into a distinct selectivity of the compounds we developed. Our top-ranked compounds are interacting with both homologous LEDGF and HRP-2 PWWP domains, yet they showed no affinity for the NSD2 PWWP1 and BRPF2 PWWP domains which belong to other PWWP domain subfamilies. Nevertheless, our developed compound series provide a strong foundation for future drug discovery targeting the LEDGF PWWP domain as they can further be explored through combinatorial chemistry. Given that the affinity of H3K36me2/3 nucleosomes to LEDGF/p75 is driven by interactions within the pocket as well as with the DNA-binding residues, we suggest that future compound development should target the latter region as well. Beyond drug discovery, our compounds can be employed to devise tool compounds to investigate the mechanism of LEDGF/p75 in epigenetic regulation.
Organizational Affiliation:
Biocrystallography, KU Leuven, Leuven, Belgium.