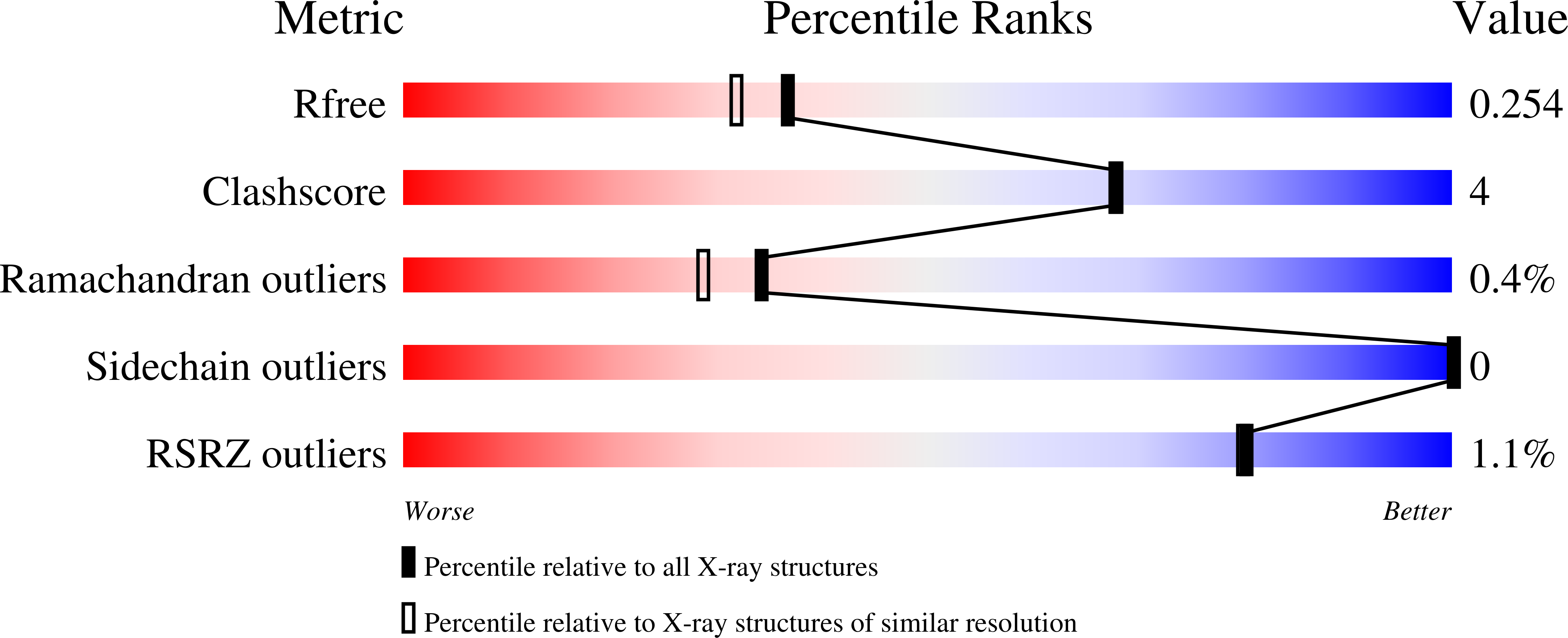
Single Residue on the WPD-Loop Affects the pH Dependency of Catalysis in Protein Tyrosine Phosphatases.
Shen, R., Crean, R.M., Johnson, S.J., Kamerlin, S.C.L., Hengge, A.C.(2021) JACS Au 1: 646-659
- PubMed: 34308419
- DOI: https://doi.org/10.1021/jacsau.1c00054
- Primary Citation of Related Structures:
7L0C, 7L0H, 7L0I, 7L0M - PubMed Abstract:
Catalysis by protein tyrosine phosphatases (PTPs) relies on the motion of a flexible protein loop (the WPD-loop) that carries a residue acting as a general acid/base catalyst during the PTP-catalyzed reaction. The orthogonal substitutions of a noncatalytic residue in the WPD-loops of YopH and PTP1B result in shifted pH-rate profiles from an altered kinetic p K a of the nucleophilic cysteine. Compared to wild type, the G352T YopH variant has a broadened pH-rate profile, similar activity at optimal pH, but significantly higher activity at low pH. Changes in the corresponding PTP1B T177G variant are more modest and in the opposite direction, with a narrowed pH profile and less activity in the most acidic range. Crystal structures of the variants show no structural perturbations but suggest an increased preference for the WPD-loop-closed conformation. Computational analysis confirms a shift in loop conformational equilibrium in favor of the closed conformation, arising from a combination of increased stability of the closed state and destabilization of the loop-open state. Simulations identify the origins of this population shift, revealing differences in the flexibility of the WPD-loop and neighboring regions. Our results demonstrate that changes to the pH dependency of catalysis by PTPs can result from small changes in amino acid composition in their WPD-loops affecting only loop dynamics and conformational equilibrium. The perturbation of kinetic p K a values of catalytic residues by nonchemical processes affords a means for nature to alter an enzyme's pH dependency by a less disruptive path than altering electrostatic networks around catalytic residues themselves.
Organizational Affiliation:
Department of Chemistry and Biochemistry, Utah State University, Logan, Utah 84322-0300, United States.