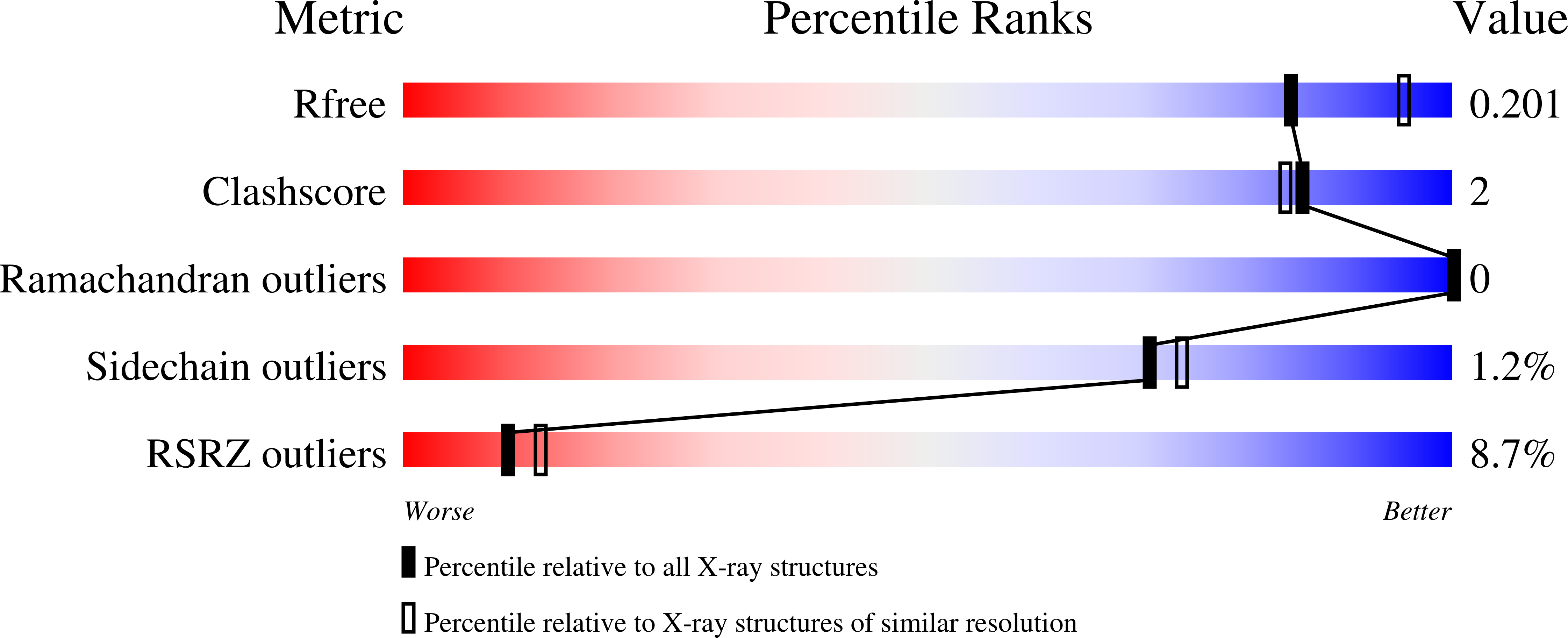
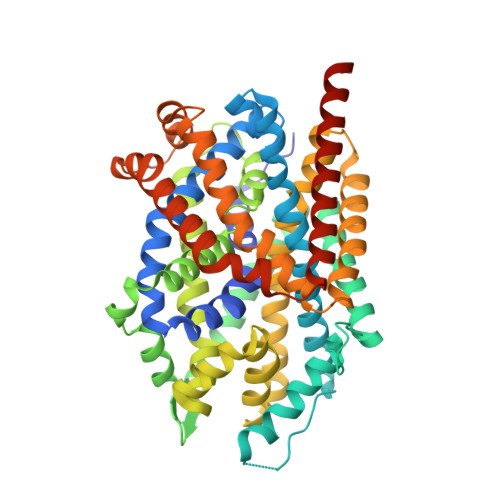
Psychomotor impairments and therapeutic implications revealed by a mutation associated with infantile Parkinsonism-Dystonia.
Aguilar, J.I., Cheng, M.H., Font, J., Schwartz, A.C., Ledwitch, K., Duran, A., Mabry, S.J., Belovich, A.N., Zhu, Y., Carter, A.M., Shi, L., Kurian, M.A., Fenollar-Ferrer, C., Meiler, J., Ryan, R.M., Mchaourab, H.S., Bahar, I., Matthies, H.J., Galli, A.(2021) Elife 10
- PubMed: 34002696
- DOI: https://doi.org/10.7554/eLife.68039
- Primary Citation of Related Structures:
7LQJ, 7LQK, 7LQL - PubMed Abstract:
Parkinson disease (PD) is a progressive, neurodegenerative disorder affecting over 6.1 million people worldwide. Although the cause of PD remains unclear, studies of highly penetrant mutations identified in early-onset familial parkinsonism have contributed to our understanding of the molecular mechanisms underlying disease pathology. Dopamine (DA) transporter (DAT) deficiency syndrome (DTDS) is a distinct type of infantile parkinsonism-dystonia that shares key clinical features with PD, including motor deficits (progressive bradykinesia, tremor, hypomimia) and altered DA neurotransmission. Here, we define structural, functional, and behavioral consequences of a Cys substitution at R445 in human DAT (hDAT R445C), identified in a patient with DTDS. We found that this R445 substitution disrupts a phylogenetically conserved intracellular (IC) network of interactions that compromise the hDAT IC gate. This is demonstrated by both Rosetta molecular modeling and fine-grained simulations using hDAT R445C, as well as EPR analysis and X-ray crystallography of the bacterial homolog leucine transporter. Notably, the disruption of this IC network of interactions supported a channel-like intermediate of hDAT and compromised hDAT function. We demonstrate that Drosophila melanogaster expressing hDAT R445C show impaired hDAT activity, which is associated with DA dysfunction in isolated brains and with abnormal behaviors monitored at high-speed time resolution. We show that hDAT R445C Drosophila exhibit motor deficits, lack of motor coordination (i.e. flight coordination) and phenotypic heterogeneity in these behaviors that is typically associated with DTDS and PD. These behaviors are linked with altered dopaminergic signaling stemming from loss of DA neurons and decreased DA availability. We rescued flight coordination with chloroquine, a lysosomal inhibitor that enhanced DAT expression in a heterologous expression system. Together, these studies shed some light on how a DTDS-linked DAT mutation underlies DA dysfunction and, possibly, clinical phenotypes shared by DTDS and PD.
Organizational Affiliation:
Department of Pharmacology, Vanderbilt University, Nashville, United States.