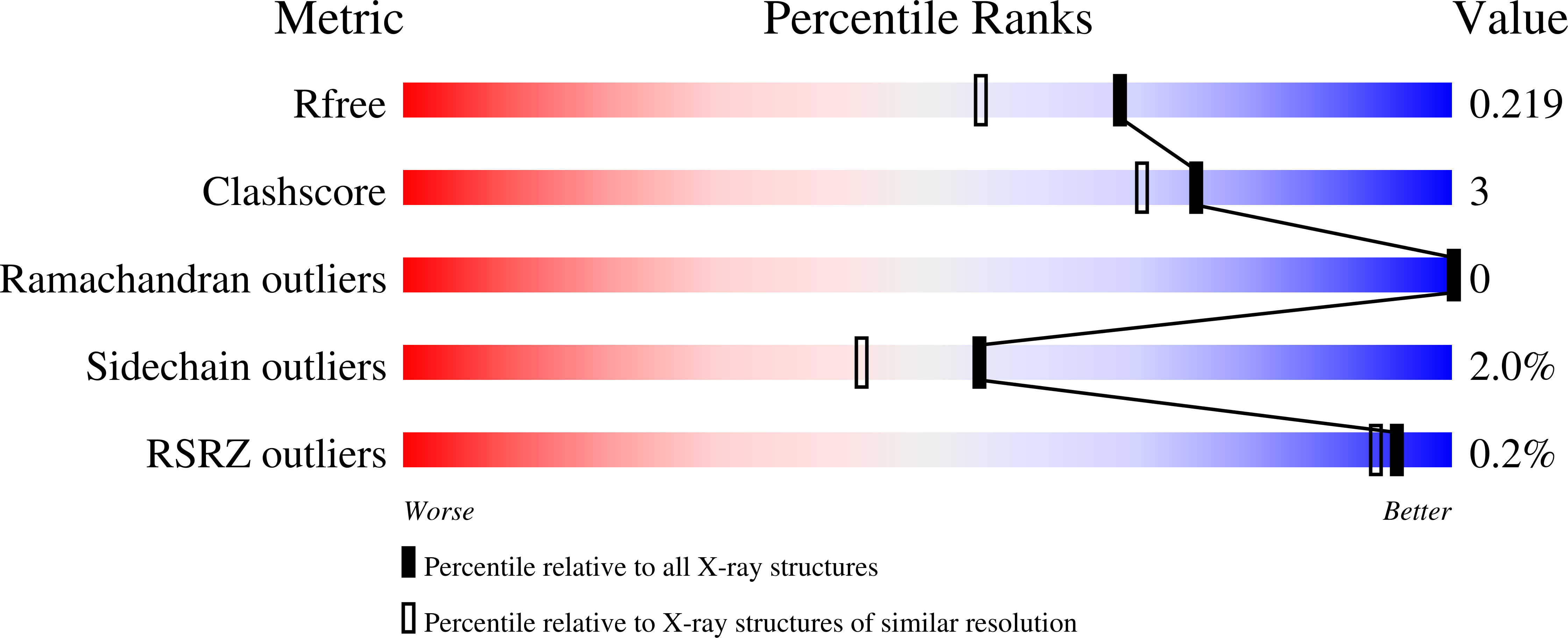
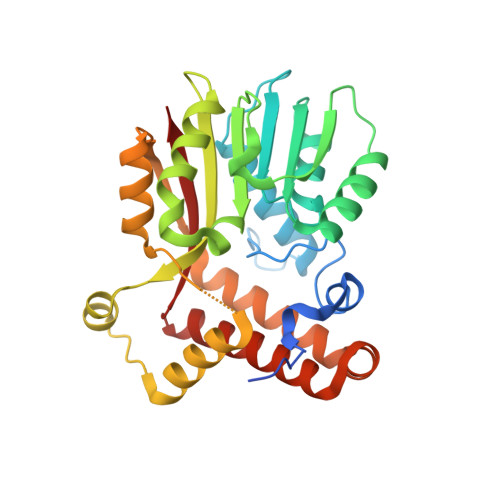
Crystal Structure of S-adenosylmethionine-dependent methyltransferase UmaA from Mycobacterium tuberculosis in complex with compound 8918
Abendroth, J., Dranow, D.M., Kahlert, G., Scarry, S.M., Grzegorzewicz, A., Jackson, M., Aube, J., Sherman, D.R., Gold, B.S., Nathan, C.F., Moritz, R.L., Lorimer, D.D., Horanyi, P.S., Edwards, T.E.To be published.