Cooperative stabilisation of 14-3-3 sigma protein-protein interactions via covalent protein modification.
Falcicchio, M., Ward, J.A., Chothia, S.Y., Basran, J., Mohindra, A., Macip, S., Roversi, P., Doveston, R.G.(2021) Chem Sci 12: 12985-12992
- PubMed: 34745529
- DOI: https://doi.org/10.1039/d1sc02120f
- Primary Citation of Related Structures:
7NFW, 7NIZ - PubMed Abstract:
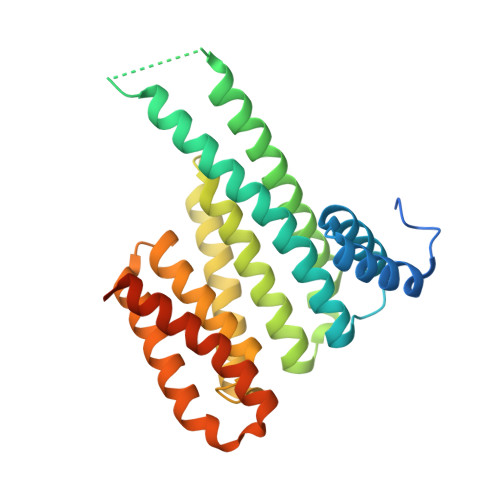
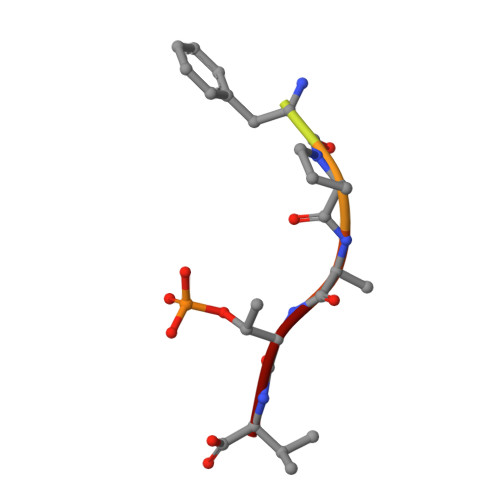
14-3-3 proteins are an important family of hub proteins that play important roles in many cellular processes via a large network of interactions with partner proteins. Many of these protein-protein interactions (PPI) are implicated in human diseases such as cancer and neurodegeneration. The stabilisation of selected 14-3-3 PPIs using drug-like 'molecular glues' is a novel therapeutic strategy with high potential. However, the examples reported to date have a number of drawbacks in terms of selectivity and potency. Here, we report that WR-1065, the active species of the approved drug amifostine, covalently modifies 14-3-3σ at an isoform-unique cysteine residue, Cys38. This modification leads to isoform-specific stabilisation of two 14-3-3σ PPIs in a manner that is cooperative with a well characterised molecular glue, fusicoccin A. Our findings reveal a novel stabilisation mechanism for 14-3-3σ, an isoform with particular involvement in cancer pathways. This mechanism can be exploited to harness the enhanced potency conveyed by covalent drug molecules and dual ligand cooperativity. This is demonstrated in two cancer cell lines whereby the cooperative behaviour of fusicoccin A and WR-1065 leads to enhanced efficacy for inducing cell death and attenuating cell growth.
Organizational Affiliation:
Leicester Institute for Structural and Chemical Biology, University of Leicester University Road Leicester LE1 7RH UK r.g.doveston@leicester.ac.uk.