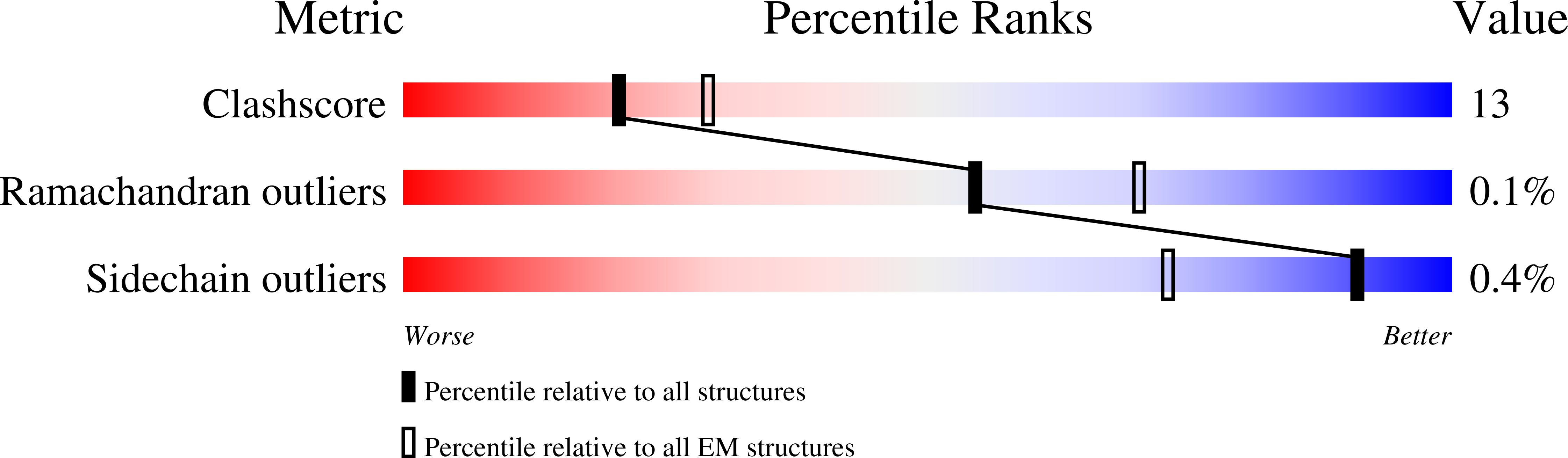Development of a universal nanobody-binding Fab module for fiducial-assisted cryo-EM studies of membrane proteins.
Bloch, J.S., Mukherjee, S., Kowal, J., Filippova, E.V., Niederer, M., Pardon, E., Steyaert, J., Kossiakoff, A.A., Locher, K.P.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 34782475
- DOI: https://doi.org/10.1073/pnas.2115435118
- Primary Citation of Related Structures:
7PHP, 7PHQ, 7PIJ, 7RTH - PubMed Abstract:
With conformation-specific nanobodies being used for a wide range of structural, biochemical, and cell biological applications, there is a demand for antigen-binding fragments (Fabs) that specifically and tightly bind these nanobodies without disturbing the nanobody-target protein interaction. Here, we describe the development of a synthetic Fab (termed NabFab) that binds the scaffold of an alpaca-derived nanobody with picomolar affinity. We demonstrate that upon complementary-determining region grafting onto this parent nanobody scaffold, nanobodies recognizing diverse target proteins and derived from llama or camel can cross-react with NabFab without loss of affinity. Using NabFab as a fiducial and size enhancer (50 kDa), we determined the high-resolution cryogenic electron microscopy (cryo-EM) structures of nanobody-bound VcNorM and ScaDMT, both small membrane proteins of ∼50 kDa. Using an additional anti-Fab nanobody further facilitated reliable initial three-dimensional structure determination from small cryo-EM test datasets. Given that NabFab is of synthetic origin, is humanized, and can be conveniently expressed in Escherichia coli in large amounts, it may be useful not only for structural biology but also for biomedical applications.
Organizational Affiliation:
Institute of Molecular Biology and Biophysics, ETH Zürich, 8093 Zürich, Switzerland.