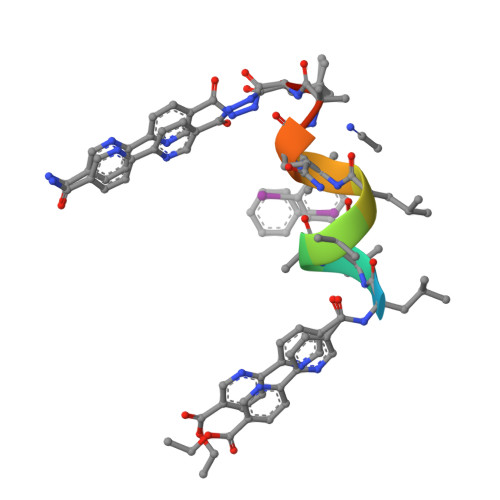
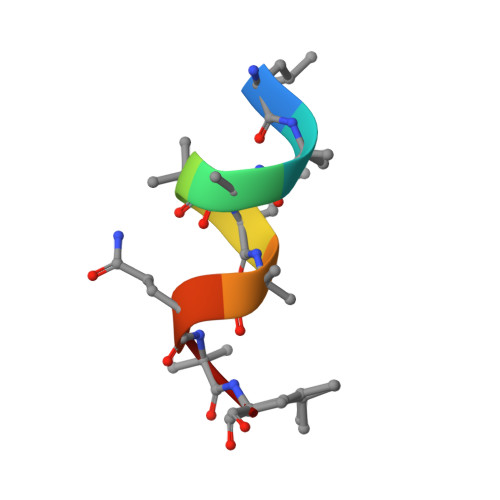
Assembly of pi-Stacking Helical Peptides into a Porous and Multivariable Proteomimetic Framework.
Heinz-Kunert, S.L., Pandya, A., Dang, V.T., Tran, P.N., Ghosh, S., McElheny, D., Santarsiero, B.D., Ren, Z., Nguyen, A.I.(2022) J Am Chem Soc 144: 7001-7009
- PubMed: 35390261
- DOI: https://doi.org/10.1021/jacs.2c02146
- Primary Citation of Related Structures:
7TLS, 7TLU, 7TM1, 7TM2, 7TMA, 7TME, 7TMH, 7TMI, 7TMJ, 7TMK, 7TML - PubMed Abstract:
The evolution of proteins from simpler, self-assembled peptides provides a powerful blueprint for the design of complex synthetic materials. Previously, peptide-metal frameworks using short sequences (≤3 residues) have shown great promise as proteomimetic materials that exhibit sophisticated capabilities. However, their development has been hindered due to few variable residues and restricted choice of side-chains that are compatible with metal ions. Herein, we developed a noncovalent strategy featuring π-stacking bipyridyl residues to assemble much longer peptides into crystalline frameworks that tolerate even previously incompatible acidic and basic functionalities and allow an unprecedented level of pore variations. Single-crystal X-ray structures are provided for all variants to guide and validate rational design. These materials exhibit hallmark proteomimetic behaviors such as guest-selective induced fit and assembly of multimetallic units. Significantly, we demonstrate facile optimization of the framework design to substantially increase affinity toward a complex organic molecule.
Organizational Affiliation:
Department of Chemistry, University of Illinois at Chicago, Chicago, Illinois 60607, United States.