Structural snapshots of V/A-ATPase reveal the rotary catalytic mechanism of rotary ATPases.
Kishikawa, J., Nakanishi, A., Nakano, A., Saeki, S., Furuta, A., Kato, T., Mistuoka, K., Yokoyama, K.(2022) Nat Commun 13: 1213-1213
- PubMed: 35260556
- DOI: https://doi.org/10.1038/s41467-022-28832-5
- Primary Citation of Related Structures:
7VAI, 7VAJ, 7VAK, 7VAL, 7VAM, 7VAN, 7VAO, 7VAP, 7VAQ, 7VAR, 7VAS, 7VAT, 7VAU, 7VAV, 7VAW, 7VAX, 7VAY, 7VB0 - PubMed Abstract:
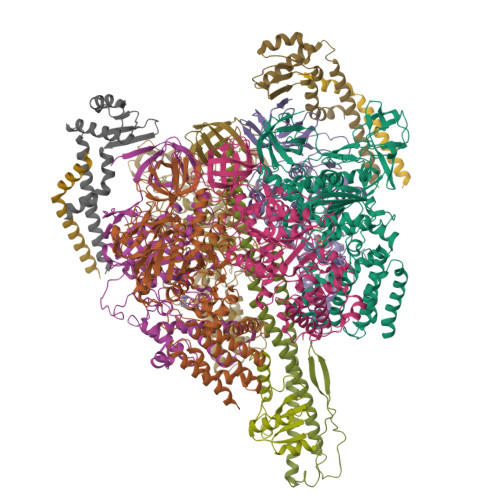
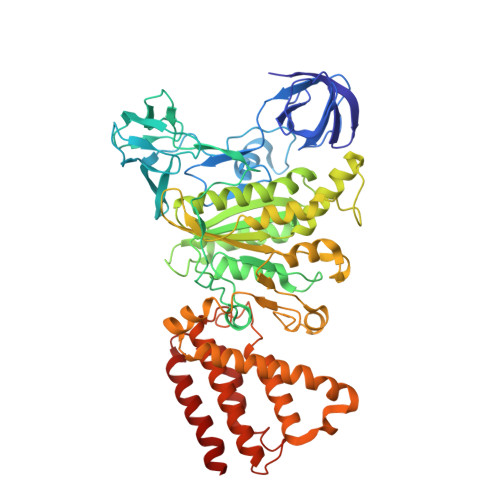
V/A-ATPase is a motor protein that shares a common rotary catalytic mechanism with F o F 1 ATP synthase. When powered by ATP hydrolysis, the V 1 domain rotates the central rotor against the A 3 B 3 hexamer, composed of three catalytic AB dimers adopting different conformations (AB open , AB semi , and AB closed ). Here, we report the atomic models of 18 catalytic intermediates of the V 1 domain of V/A-ATPase under different reaction conditions, determined by single particle cryo-EM. The models reveal that the rotor does not rotate immediately after binding of ATP to the V 1 . Instead, three events proceed simultaneously with the 120˚ rotation of the shaft: hydrolysis of ATP in AB semi , zipper movement in AB open by the binding ATP, and unzipper movement in AB closed with release of both ADP and Pi. This indicates the unidirectional rotation of V/A-ATPase by a ratchet-like mechanism owing to ATP hydrolysis in AB semi , rather than the power stroke model proposed previously for F 1 -ATPase.
Organizational Affiliation:
Department of Molecular Biosciences, Kyoto Sangyo University, Kamigamo-Motoyama, Kita-Ku, Kyoto, 603-8555, Japan.