Structure-Activity Relationships and Antileukemia Effects of the Tricyclic Benzoic Acid FTO Inhibitors.
Liu, Z., Duan, Z., Zhang, D., Xiao, P., Zhang, T., Xu, H., Wang, C.H., Rao, G.W., Gan, J., Huang, Y., Yang, C.G., Dong, Z.(2022) J Med Chem 65: 10638-10654
- PubMed: 35793358
- DOI: https://doi.org/10.1021/acs.jmedchem.2c00848
- Primary Citation of Related Structures:
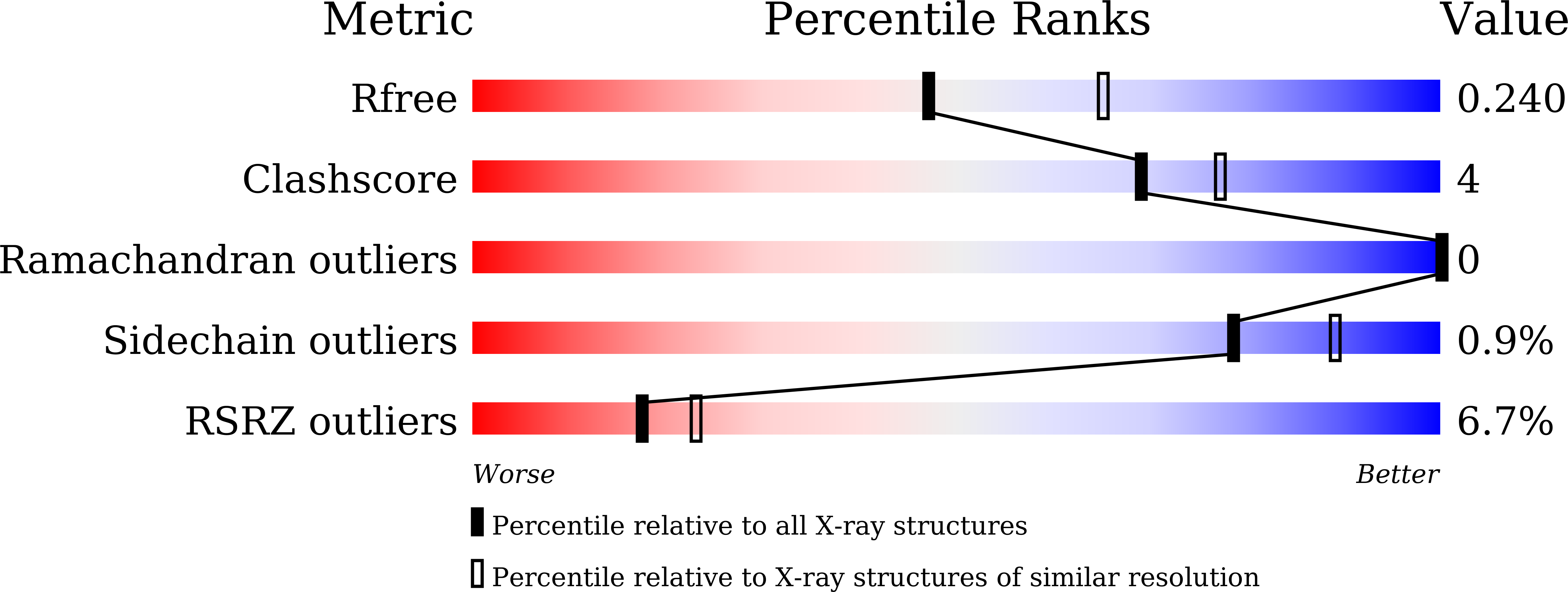
7WCV - PubMed Abstract:
The N 6 -methyladenosine (m 6 A) demethylase FTO is overexpressed in acute myeloid leukemia (AML) cells and promotes leukemogenesis. We previously developed tricyclic benzoic acid FB23 as a highly potent FTO inhibitor in vitro. However, it showed a moderate antiproliferative effect on AML cells. In this work, we performed a structure-activity relationship study of tricyclic benzoic acids as FTO inhibitors. The analog 13a exhibited excellent inhibitory effects on FTO similar to that of FB23 in vitro. In contrast to FB23, 13a exerted a strong antiproliferative effect on AML cells. Like FTO knock down, 13a upregulated ASB2 and RARA expression and increased the protein abundance while it downregulated MYC expression and decreased MYC protein abundance. These genes are key FTO targets in AML cells. Finally, 13a treatment improved the survival rate of MONOMAC6-transplanted NSG mice. Collectively, our data suggest that targeting FTO with tricyclic benzoic acid inhibitors may be a potential strategy for treating AML.
Organizational Affiliation:
State Key Laboratory of Drug Research, Centre for the Chemical Biology, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, China.