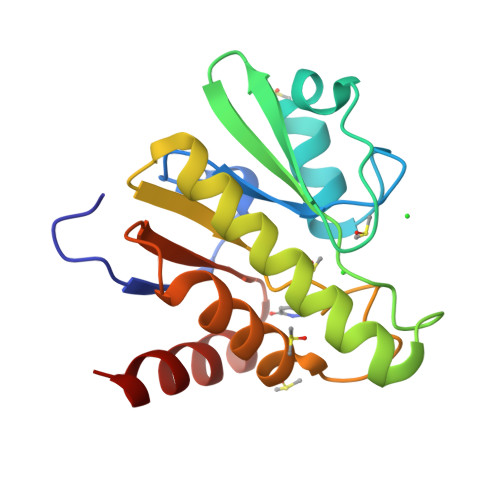
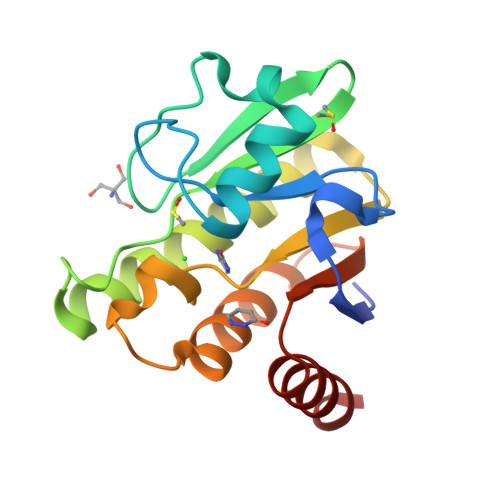
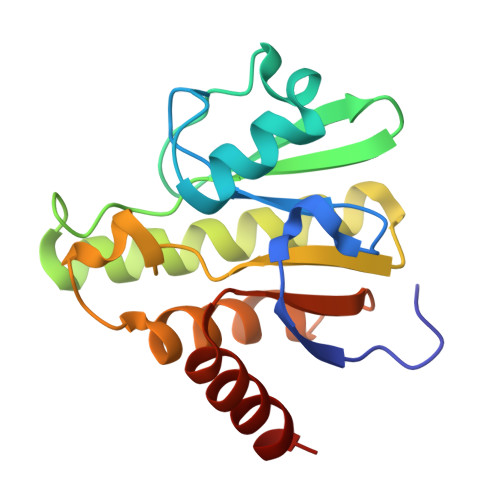
MPB (Subject of Investigation/LOI) Query on MPB Download Ideal Coordinates CCD File L [auth A] 4-HYDROXY-BENZOIC ACID METHYL ESTER 8 H8 O3 TRS Query on TRS Download Ideal Coordinates CCD File AA [auth D], 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL 4 H12 N O3 W3G (Subject of Investigation/LOI) Query on W3G Download Ideal Coordinates CCD File GA [auth D], pyridazin-3(2H)-one 4 H4 N2 OXJV (Subject of Investigation/LOI) Query on XJV Download Ideal Coordinates CCD File FA [auth D], imidazolidin-2-one 3 H6 N2 ODMS Query on DMS Download Ideal Coordinates CCD File BA [auth D]
BA [auth D],
DIMETHYL SULFOXIDE 2 H6 O SCL Query on CL Download Ideal Coordinates CCD File EA [auth D]
EA [auth D],
CHLORIDE ION