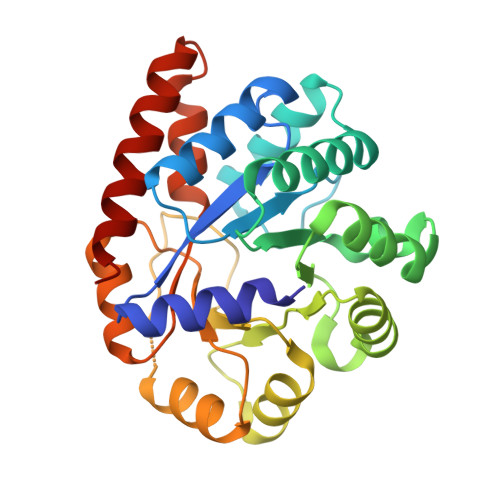
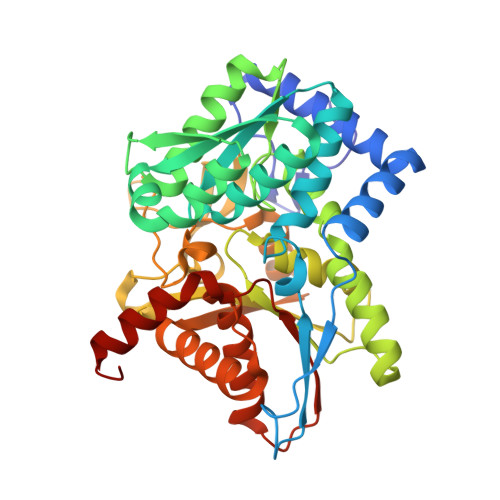
The internal aldimine form of the wild-type Salmonella Typhimurium Tryptophan Synthase in complex with N-(4'-trifluoromethoxybenzoyl)-2-amino-1-ethylphosphate (F6F) inhibitor at the alpha-and beta-site, sodium ion at the metal coordination site, and another F6F molecule at the enzyme beta-site at 1.55 Angstrom resolution. One of the beta-Q114 rotamer conformations allows a hydrogen bond to form with the PLP oxygen at the position 3 in the ring.
Hilario, E., Dunn, M.F., Mueller, L.J.To be published.