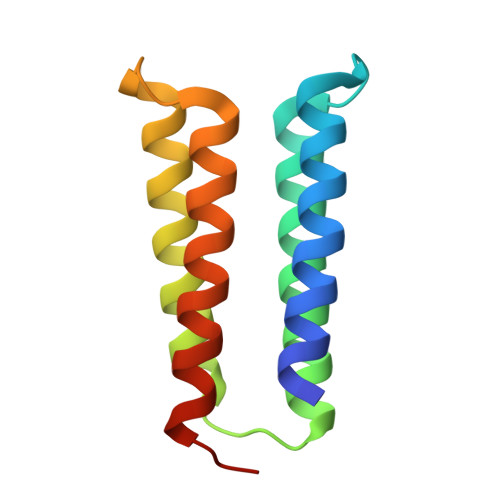
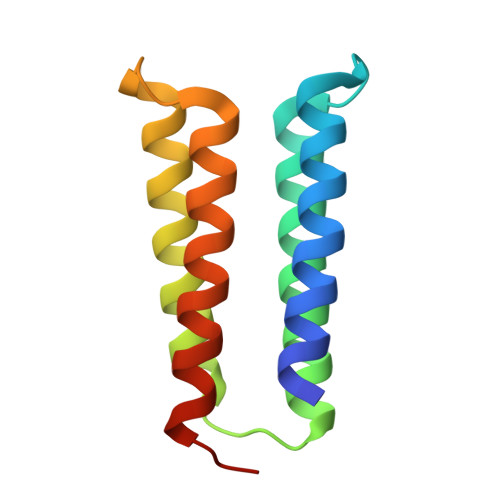
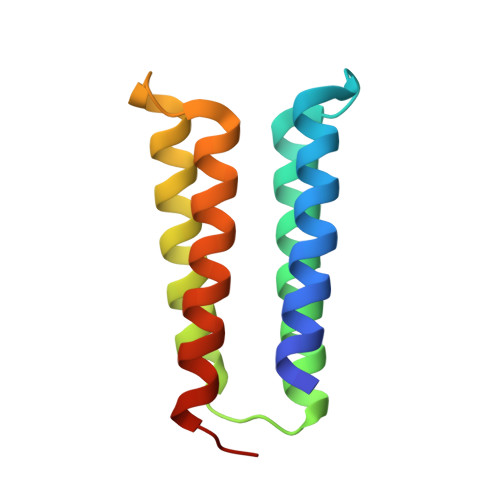
Hallucination of closed repeat proteins containing central pockets.
An, L., Hicks, D.R., Zorine, D., Dauparas, J., Wicky, B.I.M., Milles, L.F., Courbet, A., Bera, A.K., Nguyen, H., Kang, A., Carter, L., Baker, D.(2023) Nat Struct Mol Biol 30: 1755-1760
- PubMed: 37770718
- DOI: https://doi.org/10.1038/s41594-023-01112-6
- Primary Citation of Related Structures:
8FJE, 8FJF, 8FJG - PubMed Abstract:
In pseudocyclic proteins, such as TIM barrels, β barrels, and some helical transmembrane channels, a single subunit is repeated in a cyclic pattern, giving rise to a central cavity that can serve as a pocket for ligand binding or enzymatic activity. Inspired by these proteins, we devised a deep-learning-based approach to broadly exploring the space of closed repeat proteins starting from only a specification of the repeat number and length. Biophysical data for 38 structurally diverse pseudocyclic designs produced in Escherichia coli are consistent with the design models, and the three crystal structures we were able to obtain are very close to the designed structures. Docking studies suggest the diversity of folds and central pockets provide effective starting points for designing small-molecule binders and enzymes.
- Department of Biochemistry, University of Washington, Seattle, WA, USA. linnaan@uw.edu.
Organizational Affiliation: