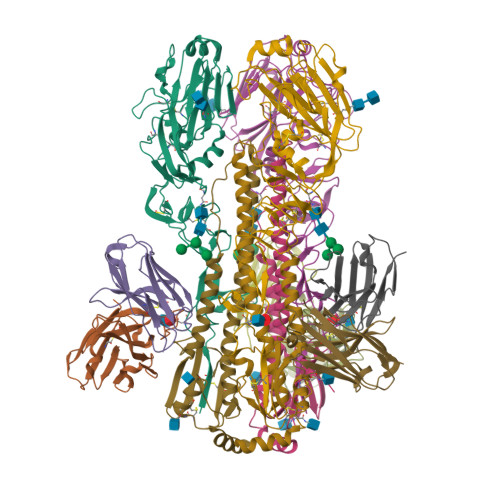
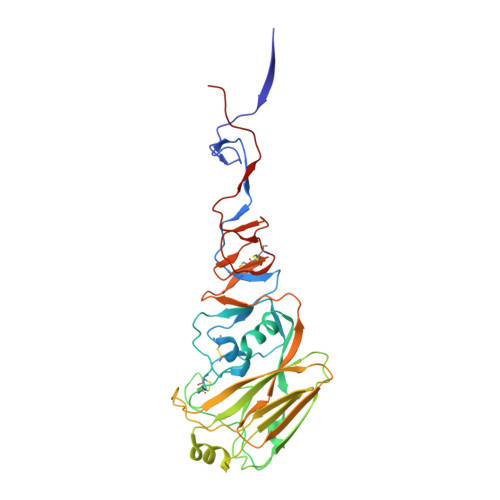
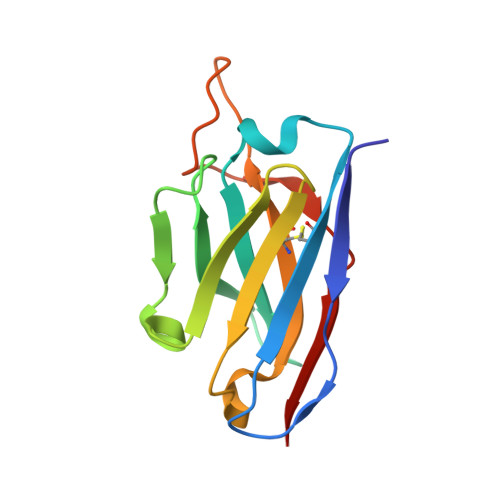
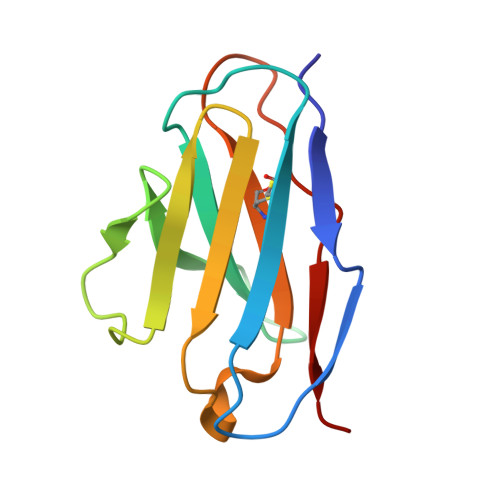
Structural basis for a human broadly neutralizing influenza A hemagglutinin stem-specific antibody including H17/18 subtypes.
Chen, Y., Wang, F., Yin, L., Jiang, H., Lu, X., Bi, Y., Zhang, W., Shi, Y., Burioni, R., Tong, Z., Song, H., Qi, J., Gao, G.F.(2022) Nat Commun 13: 7603-7603
- PubMed: 36494358
- DOI: https://doi.org/10.1038/s41467-022-35236-y
- Primary Citation of Related Structures:
7WVD, 7WVG, 7WVI, 8GV4, 8GV5, 8GV6, 8GV7 - PubMed Abstract:
Influenza infection continues are a persistent threat to public health. The identification and characterization of human broadly neutralizing antibodies can facilitate the development of antibody drugs and the design of universal influenza vaccines. Here, we present structural information for the human antibody PN-SIA28's heterosubtypic binding of hemagglutinin (HA) from circulating and emerging potential influenza A viruses (IAVs). Aside from group 1 and 2 conventional IAV HAs, PN-SIA28 also inhibits membrane fusion mediated by bat-origin H17 and H18 HAs. Crystallographic analyses of Fab alone or in complex with H1, H14, and H18 HA proteins reveal that PN-SIA28 binds to a highly conserved epitope in the fusion domain of different HAs, with the same CDRHs but different CDRLs for different HAs tested, distinguishing it from other structurally characterized anti-stem antibodies. The binding characteristics of PN-SIA28 provides information to support the design of increasingly potent engineered antibodies, antiviral drugs, and/or universal influenza vaccines.
Organizational Affiliation:
CAS Key Laboratory of Pathogen Microbiology and Immunology, Institute of Microbiology, Chinese Academy of Sciences, Beijing, 100101, China.