Molecular basis of SAP05-mediated ubiquitin-independent proteasomal degradation of transcription factors.
Yan, X., Yuan, X., Lv, J., Zhang, B., Huang, Y., Li, Q., Ma, J., Li, Y., Wang, X., Li, Y., Yu, Y., Liu, Q., Liu, T., Mi, W., Dong, C.(2024) Nat Commun 15: 1170-1170
- PubMed: 38326322
- DOI: https://doi.org/10.1038/s41467-024-45521-7
- Primary Citation of Related Structures:
8J48, 8J49, 8J4A, 8J4B - PubMed Abstract:
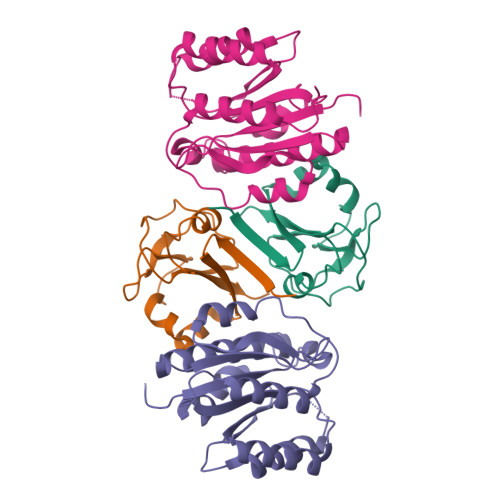
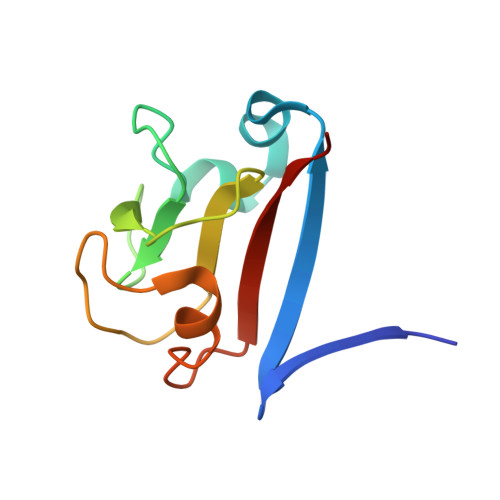
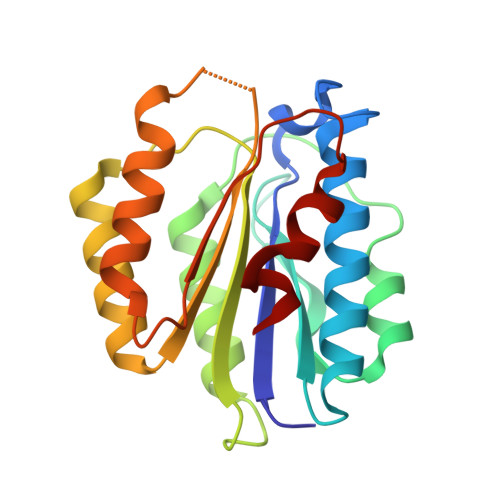
SAP05, a secreted effector by the obligate parasitic bacteria phytoplasma, bridges host SPL and GATA transcription factors (TFs) to the 26 S proteasome subunit RPN10 for ubiquitination-independent degradation. Here, we report the crystal structures of SAP05 in complex with SPL5, GATA18 and RPN10, which provide detailed insights into the protein-protein interactions involving SAP05. SAP05 employs two opposing lobes with an acidic path and a hydrophobic path to contact TFs and RPN10, respectively. Our crystal structures, in conjunction with mutagenesis and degradation assays, reveal that SAP05 targets plant GATAs but not animal GATAs dependent on their direct salt-bridged electrostatic interactions. Additionally, SAP05 hijacks plant RPN10 but not animal RPN10 due to structural steric hindrance and the key hydrophobic interactions. This study provides valuable molecular-level information into the modulation of host proteins to prevent insect-borne diseases.
Organizational Affiliation:
The Province and Ministry Co-sponsored Collaborative Innovation Center for Medical Epigenetics, Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), School of Basic Medical Sciences, Tianjin Medical University, Tianjin, 300070, China.