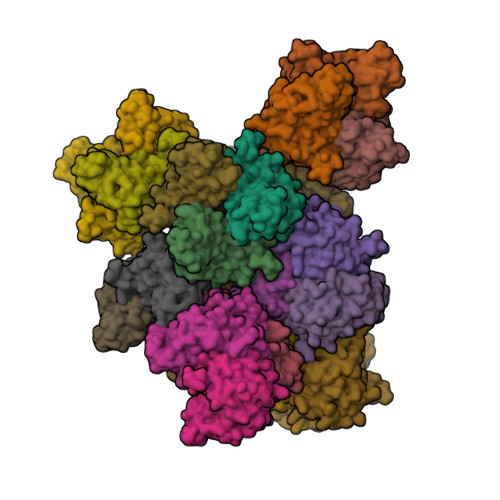
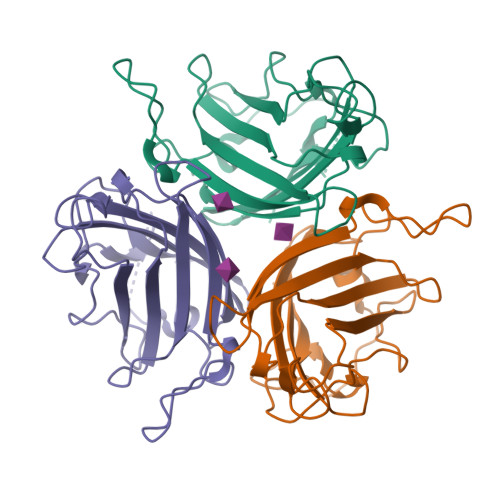
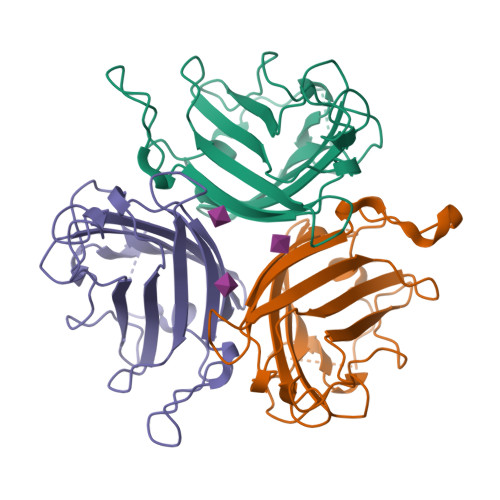
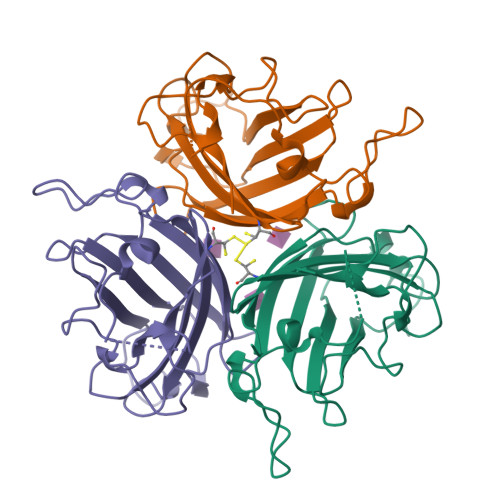
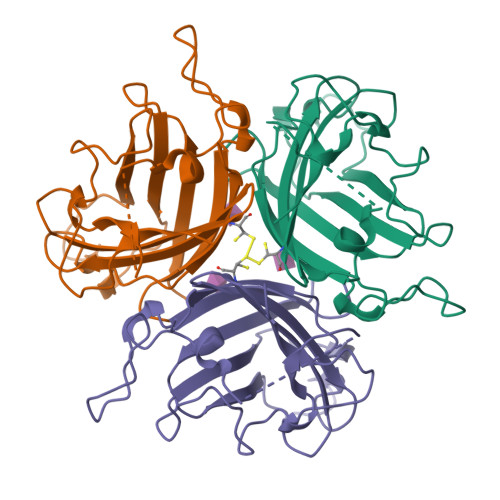
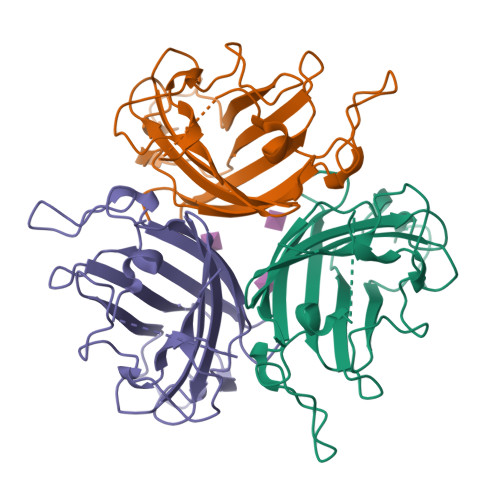
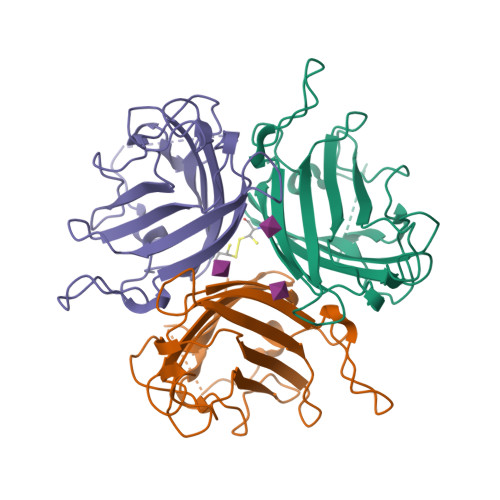
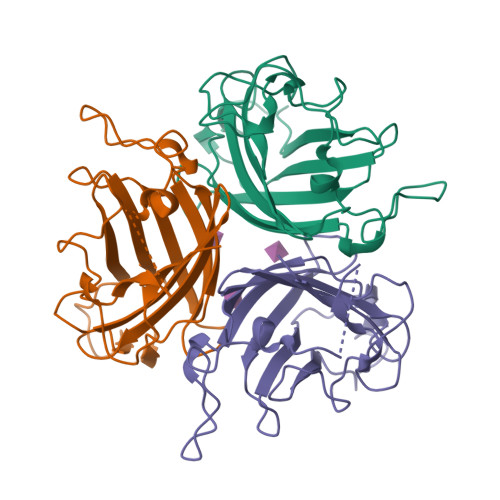
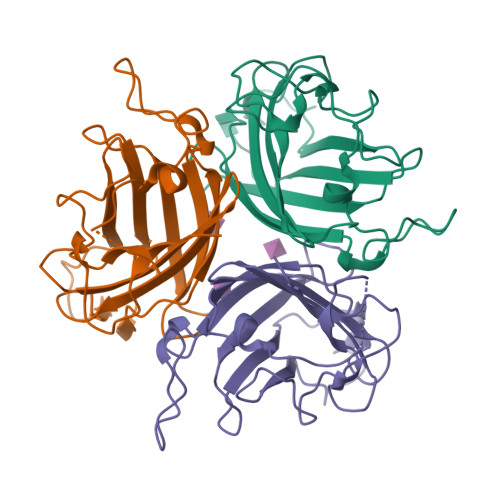
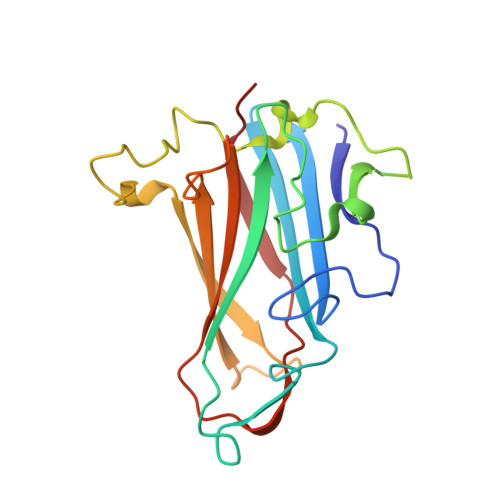
Broad sialic acid usage amongst species D human adenovirus.
Mundy, R.M., Baker, A.T., Bates, E.A., Cunliffe, T.G., Teijeira-Crespo, A., Moses, E., Rizkallah, P.J., Parker, A.L.(2023) Npj Viruses 1: 1-1
- PubMed: 38665237
- DOI: https://doi.org/10.1038/s44298-023-00001-5
- Primary Citation of Related Structures:
8OFP, 8OFQ, 8OFR, 8OFS, 8OFT, 8OFU, 8OFV - PubMed Abstract:
Human adenoviruses (HAdV) are widespread pathogens causing usually mild infections. The Species D (HAdV-D) cause gastrointestinal tract infections and epidemic keratoconjunctivitis (EKC). Despite being significant pathogens, knowledge around HAdV-D mechanism of cell infection is lacking. Sialic acid (SA) usage has been proposed as a cell infection mechanism for EKC causing HAdV-D. Here we highlight an important role for SA engagement by many HAdV-D. We provide apo state crystal structures of 7 previously undetermined HAdV-D fiber-knob proteins, and structures of HAdV-D25, D29, D30 and D53 fiber-knob proteins in complex with SA. Biologically, we demonstrate that removal of cell surface SA reduced infectivity of HAdV-C5 vectors pseudotyped with HAdV-D fiber-knob proteins, whilst engagement of the classical HAdV receptor CAR was variable. Our data indicates variable usage of SA and CAR across HAdV-D. Better defining these interactions will enable improved development of antivirals and engineering of the viruses into refined therapeutic vectors.
Organizational Affiliation:
Division of Cancer & Genetics, Cardiff University School of Medicine, Cardiff, CF14 4XN UK.