HsNMT1 in complex with both MyrCoA and Ac-D-ORN-SFSKPR inhibitor peptide
Dian, C., Giglione, C., Meinnel, T., Riviere, F., Dutheil, F.R., Monassa, P.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Glycylpeptide N-tetradecanoyltransferase 1 | 402 | Homo sapiens | Mutation(s): 0 Gene Names: NMT1, NMT EC: 2.3.1.97 (PDB Primary Data), 2.3.1 (UniProt) | 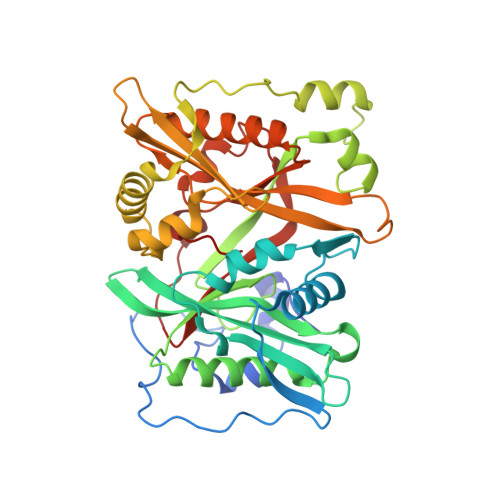 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P30419 (Homo sapiens) Explore P30419 Go to UniProtKB: P30419 | |||||
PHAROS: P30419 GTEx: ENSG00000136448 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P30419 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ACE-ORD-SER-PHE-SER-LYS-PRO-ARG | C [auth I], D [auth E] | 8 | synthetic construct | Mutation(s): 0 | 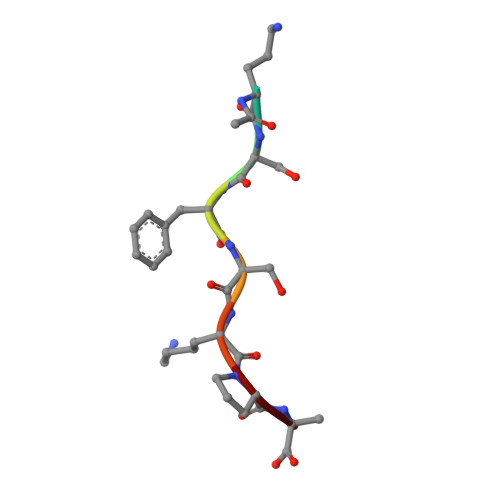 |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| MYA (Subject of Investigation/LOI) Query on MYA | G [auth A], K [auth B] | TETRADECANOYL-COA C35 H62 N7 O17 P3 S DUAFKXOFBZQTQE-QSGBVPJFSA-N |  | ||
| GOL Query on GOL | E [auth A], H [auth A], I [auth B], J [auth B], L [auth B] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| CL Query on CL | F [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 79.815 | α = 90 |
| b = 179.102 | β = 90 |
| c = 58.367 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Agence Nationale de la Recherche (ANR) | France | ANR-20-CE44-0013 |
| Agence Nationale de la Recherche (ANR) | France | ANR-2010-BLAN-1611-01 |
| Fondation ARC | France | ARCPJA32020060002137 |
| French Infrastructure for Integrated Structural Biology (FRISBI) | France | ANR-10-INSB-05-01 |