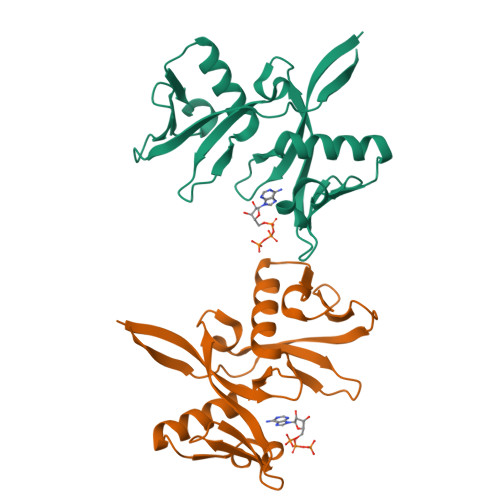
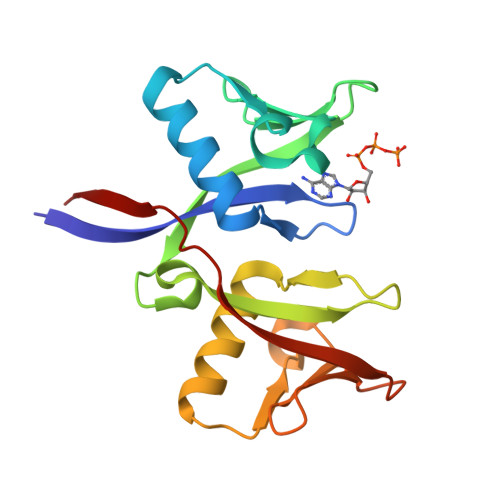
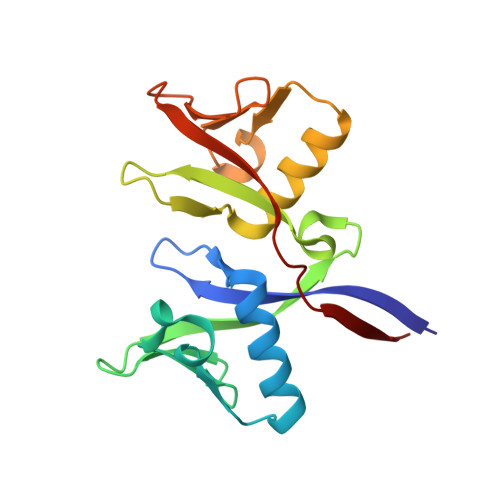
A ligand discovery toolbox for the WWE domain family of human E3 ligases.
Munzker, L., Kimani, S.W., Fowkes, M.M., Dong, A., Zheng, H., Li, Y., Dasovich, M., Zak, K.M., Leung, A.K.L., Elkins, J.M., Kessler, D., Arrowsmith, C.H., Halabelian, L., Bottcher, J.(2024) Commun Biol 7: 901-901
- PubMed: 39048679
- DOI: https://doi.org/10.1038/s42003-024-06584-w
- Primary Citation of Related Structures:
8R5N, 8R6A, 8R6B, 8R7O, 8RD0, 8RD1, 8RD7 - PubMed Abstract:
The WWE domain is a relatively under-researched domain found in twelve human proteins and characterized by a conserved tryptophan-tryptophan-glutamate (WWE) sequence motif. Six of these WWE domain-containing proteins also contain domains with E3 ubiquitin ligase activity. The general recognition of poly-ADP-ribosylated substrates by WWE domains suggests a potential avenue for development of Proteolysis-Targeting Chimeras (PROTACs). Here, we present novel crystal structures of the HUWE1, TRIP12, and DTX1 WWE domains in complex with PAR building blocks and their analogs, thus enabling a comprehensive analysis of the PAR binding site structural diversity. Furthermore, we introduce a versatile toolbox of biophysical and biochemical assays for the discovery and characterization of novel WWE domain binders, including fluorescence polarization-based PAR binding and displacement assays, 15 N-NMR-based binding affinity assays and 19 F-NMR-based competition assays. Through these assays, we have characterized the binding of monomeric iso-ADP-ribose (iso-ADPr) and its nucleotide analogs with the aforementioned WWE proteins. Finally, we have utilized the assay toolbox to screen a small molecule fragment library leading to the successful discovery of novel ligands targeting the HUWE1 WWE domain.
Organizational Affiliation:
Boehringer Ingelheim RCV GmbH & Co KG, Vienna, Austria.