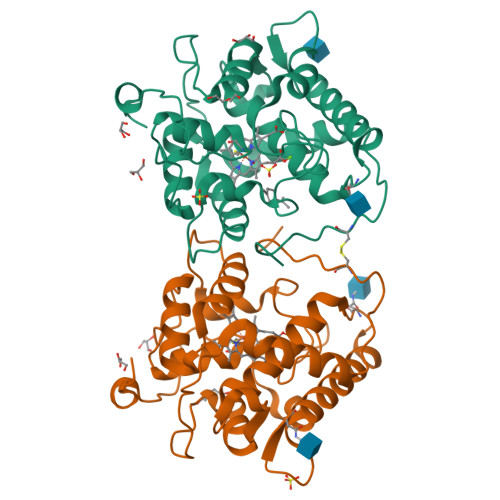
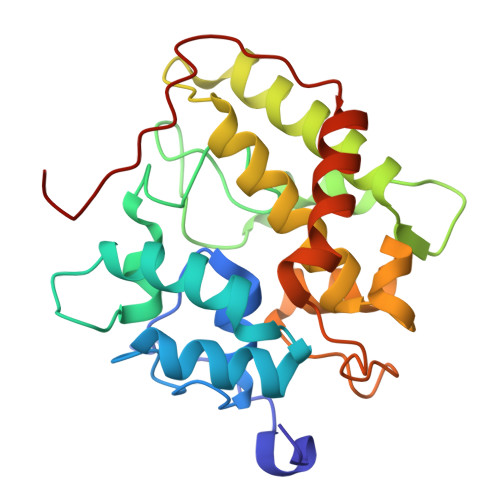
Structural Insights and Reaction Profile of a New Unspecific Peroxygenase from Marasmius wettsteinii Produced in a Tandem-Yeast Expression System.
Sanchez-Moreno, I., Fernandez-Garcia, A., Mateljak, I., Gomez de Santos, P., Hofrichter, M., Kellner, H., Sanz-Aparicio, J., Alcalde, M.(2024) ACS Chem Biol 19: 2240-2253
- PubMed: 39367827
- DOI: https://doi.org/10.1021/acschembio.4c00504
- Primary Citation of Related Structures:
8RNJ, 8RNK, 8RNL, 8RNM, 8RNN, 8RNO, 8RNP, 8RNQ, 8RNR - PubMed Abstract:
Fungal unspecific peroxygenases (UPOs) are gaining momentum in synthetic chemistry. Of special interest is the UPO from Marasmius rotula ( Mro UPO), which shows an exclusive repertoire of oxyfunctionalizations, including the terminal hydroxylation of alkanes, the α-oxidation of fatty acids and the C-C cleavage of corticosteroids. However, the lack of heterologous expression systems to perform directed evolution has impeded its engineering for practical applications. Here, we introduce a close ortholog of Mro UPO, a UPO gene from Marasmius wettsteinii ( Mwe UPO-1), that has a similar reaction profile to Mro UPO and for which we have set up a directed evolution platform based on tandem-yeast expression. Recombinant Mwe UPO-1 was produced at high titers in the bioreactor (0.7 g/L) and characterized at the biochemical and atomic levels. The conjunction of soaking crystallographic experiments at a resolution up to 1.6 Å together with the analysis of reaction patterns sheds light on the substrate preferences of this promiscuous biocatalyst.
Organizational Affiliation:
Department of Biocatalysis, Institute of Catalysis, CSIC, 28049 Madrid, Spain.