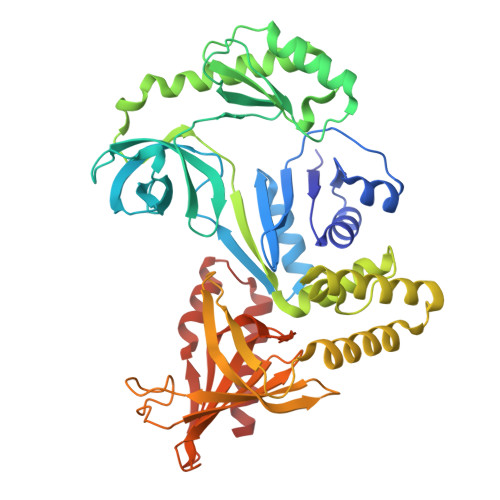
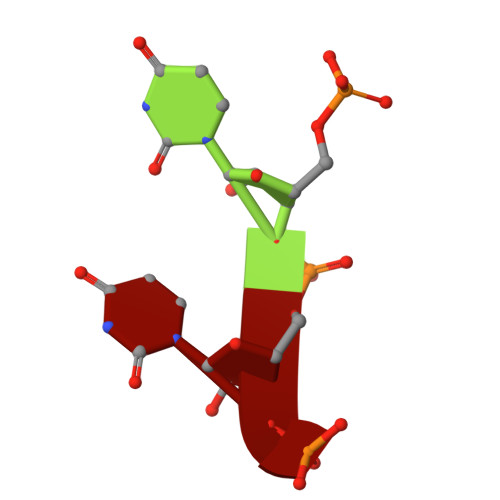
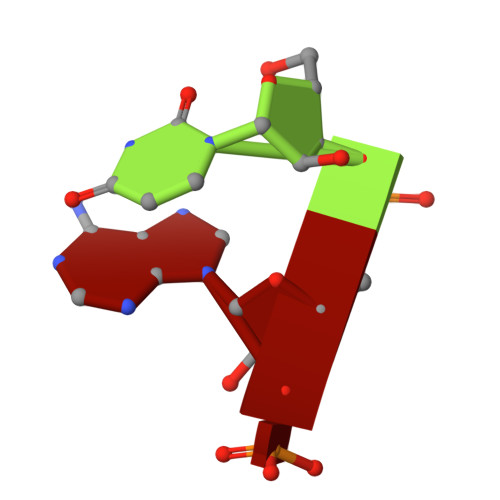
RNase W, a conserved ribonuclease family with a novel active site.
Vayssieres, M., Juttner, M., Haas, K., Ancelin, A., Marchfelder, A., Leulliot, N., Ferreira-Cerca, S., Blaud, M.(2024) Nucleic Acids Res
- PubMed: 39445822
- DOI: https://doi.org/10.1093/nar/gkae907
- Primary Citation of Related Structures:
8RZA, 8RZF - PubMed Abstract:
Ribosome biogenesis is a complex process requiring multiple precursor ribosomal RNA (rRNA) cleavage steps. In archaea, the full set of ribonucleases (RNases) involved in rRNA processing remains to be discovered. A previous study suggested that FAU-1, a conserved protein containing an RNase G/E-like protein domain fused to a domain of unknown function (DUF402), acts as an RNase in archaea. However, the molecular basis of this activity remained so far elusive. Here, we report two X-ray crystallographic structures of RNase G/E-like-DUF402 hybrid proteins from Pyrococcus furiosus and Sulfolobus acidocaldarius, at 2.1 and 2.0 Å, respectively. The structures highlight a structural homology with the 5' RNA recognition domain of Escherichia coli RNase E but no homology with other known catalytic nuclease domains. Surprisingly, we demonstrate that the C-terminal domain of this hybrid protein, annotated as a putative diphosphatase domain, harbors the RNase activity. Our functional analysis also supports a model by which the RNase G/E-like domain acts as a regulatory subunit of the RNase activity. Finally, in vivo experiments in Haloferax volcanii suggest that this RNase participates in the maturation of pre-16S rRNA. Together, our study defines a new RNase family, which we termed the RNase W family, as the first archaea-specific contributor to archaeal ribosome biogenesis.
Organizational Affiliation:
Université Paris Cité, CNRS, CiTCoM, 4 avenue de l'Observatoire, F-75006 Paris, France.