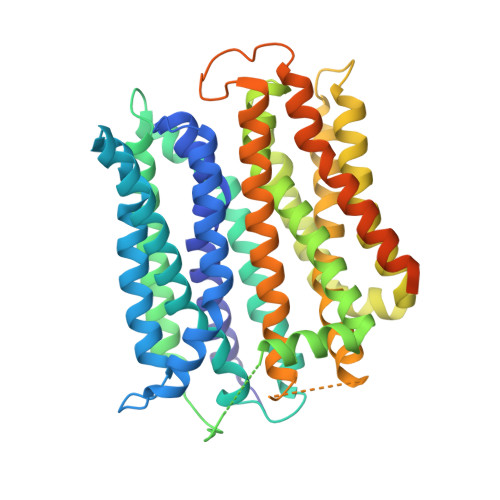
Mobile barrier mechanisms for Na + -coupled symport in an MFS sugar transporter.
Hariharan, P., Shi, Y., Katsube, S., Willibal, K., Burrows, N.D., Mitchell, P., Bakhtiiari, A., Stanfield, S., Pardon, E., Kaback, H.R., Liang, R., Steyaert, J., Viner, R., Guan, L.(2024) Elife 12
- PubMed: 38381130
- DOI: https://doi.org/10.7554/eLife.92462
- Primary Citation of Related Structures:
8T60 - PubMed Abstract:
While many 3D structures of cation-coupled transporters have been determined, the mechanistic details governing the obligatory coupling and functional regulations still remain elusive. The bacterial melibiose transporter (MelB) is a prototype of major facilitator superfamily transporters. With a conformation-selective nanobody, we determined a low-sugar affinity inward-facing Na + -bound cryoEM structure. The available outward-facing sugar-bound structures showed that the N- and C-terminal residues of the inner barrier contribute to the sugar selectivity. The inward-open conformation shows that the sugar selectivity pocket is also broken when the inner barrier is broken. Isothermal titration calorimetry measurements revealed that this inward-facing conformation trapped by this nanobody exhibited a greatly decreased sugar-binding affinity, suggesting the mechanisms for substrate intracellular release and accumulation. While the inner/outer barrier shift directly regulates the sugar-binding affinity, it has little or no effect on the cation binding, which is supported by molecular dynamics simulations. Furthermore, the hydron/deuterium exchange mass spectrometry analyses allowed us to identify dynamic regions; some regions are involved in the functionally important inner barrier-specific salt-bridge network, which indicates their critical roles in the barrier switching mechanisms for transport. These complementary results provided structural and dynamic insights into the mobile barrier mechanism for cation-coupled symport.
Organizational Affiliation:
Department of Cell Physiology and Molecular Biophysics, Center for Membrane Protein Research, Texas Tech University Health Sciences Center, School of Medicine, Lubbock, United States.