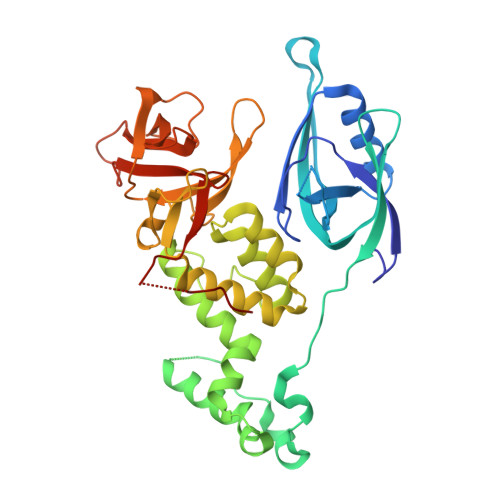
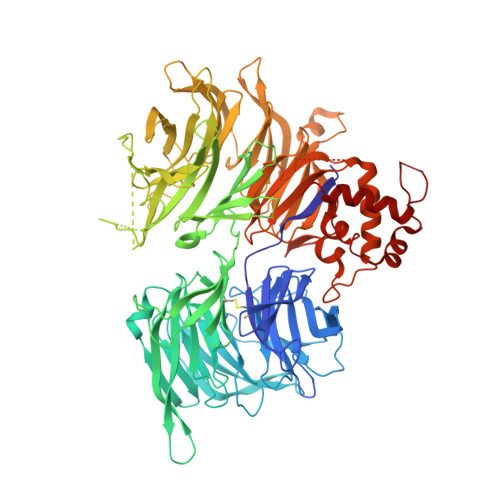
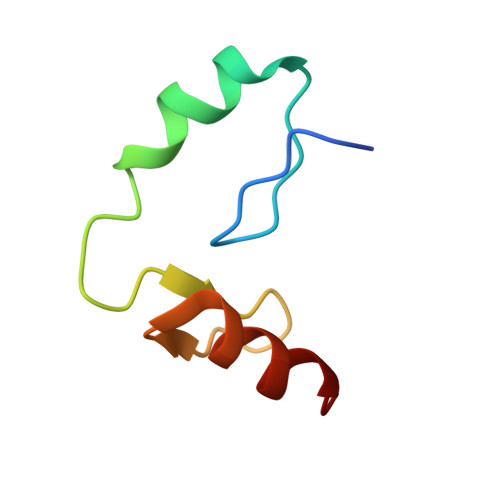
Structural and biophysical comparisons of the pomalidomide- and CC-220-induced interactions of SALL4 with cereblon.
Ma, X., Leon, B., Ornelas, E., Dovala, D., Tandeske, L., Luu, C., Pardee, G., Widger, S., Solomon, J.M., Beckwith, R.E.J., Moser, H., Clifton, M.C., Wartchow, C.A.(2023) Sci Rep 13: 22088-22088
- PubMed: 38086859
- DOI: https://doi.org/10.1038/s41598-023-48606-3
- Primary Citation of Related Structures:
8U15, 8U16, 8U17 - PubMed Abstract:
The design of cereblon-binding molecular glues (MGs) that selectively recruit a desired protein while excluding teratogenic SALL4 is an area of significant interest when designing therapeutic agents. Previous studies show that SALL4 is degraded in the presence of IKZF1 degraders pomalidomide, and to a lesser extent by CC-220. To expand our understanding of the molecular basis for the interaction of SALL4 with cereblon, we performed biophysical and structural studies demonstrating that SALL4 zinc finger domains one and two (ZF1-2) interact with cereblon (CRBN) in a unique manner. ZF1 interacts with the N-terminal domain of cereblon and ZF2 binds as expected in the C-terminal IMiD-binding domain. Both ZF1 and ZF2 contribute to the potency of the interaction of ZF1-2 with CRBN:MG complexes and the affinities of SALL4 ZF1-2 for the cereblon:CC-220 complex are less potent than for the corresponding pomalidomide complex. Structural analysis provides a rationale for understanding the reduced affinity of SALL4 for cereblon in the presence of CC-220, which engages both ZF1 and ZF2. These studies further our understanding of the molecular glue-mediated interactions of zinc finger-based proteins with cereblon and may provide structural tools for the prospective design of compounds with reduced binding and degradation of SALL4.
Organizational Affiliation:
Global Discovery Chemistry, Novartis Biomedical Research, Emeryville, CA, 94608, USA.