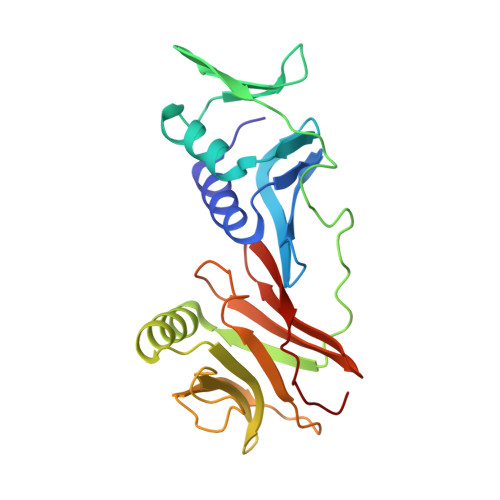
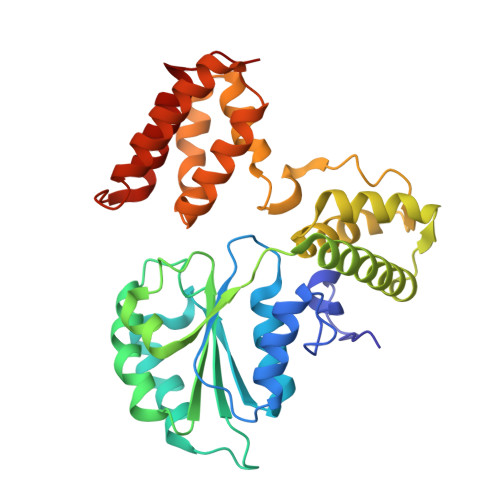
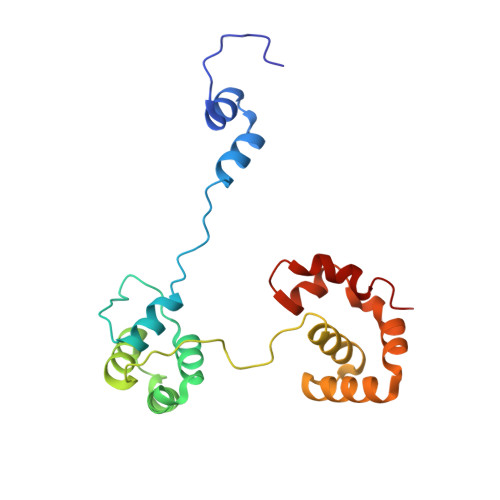
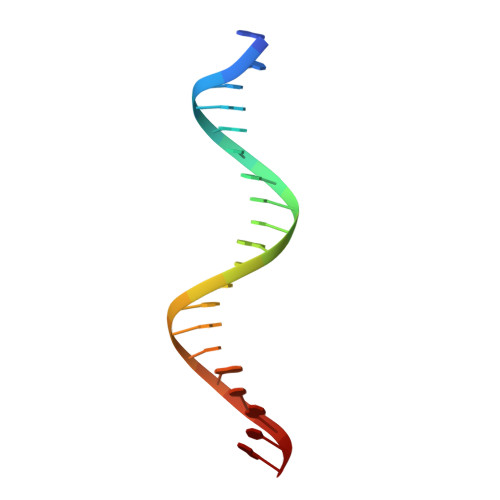
Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Marcus, K., Huang, Y., Subramanian, S., Gee, C.L., Gorday, K., Ghaffari-Kashani, S., Luo, X.R., Zheng, L., O'Donnell, M., Subramaniam, S., Kuriyan, J.(2024) Nat Struct Mol Biol 31: 424-435
- PubMed: 38177685
- DOI: https://doi.org/10.1038/s41594-023-01177-3
- Primary Citation of Related Structures:
8UH7, 8UK9, 8UNF, 8UNH - PubMed Abstract:
Clamp loaders are AAA+ ATPases that facilitate high-speed DNA replication. In eukaryotic and bacteriophage clamp loaders, ATP hydrolysis requires interactions between aspartate residues in one protomer, present in conserved 'DEAD-box' motifs, and arginine residues in adjacent protomers. We show that functional defects resulting from a DEAD-box mutation in the T4 bacteriophage clamp loader can be compensated by widely distributed single mutations in the ATPase domain. Using cryo-EM, we discovered an unsuspected inactive conformation of the clamp loader, in which DNA binding is blocked and the catalytic sites are disassembled. Mutations that restore function map to regions of conformational change upon activation, suggesting that these mutations may increase DNA affinity by altering the energetic balance between inactive and active states. Our results show that there are extensive opportunities for evolution to improve catalytic efficiency when an inactive intermediate is involved.
Organizational Affiliation:
Department of Biochemistry, Vanderbilt University School of Medicine, Nashville, TN, USA.