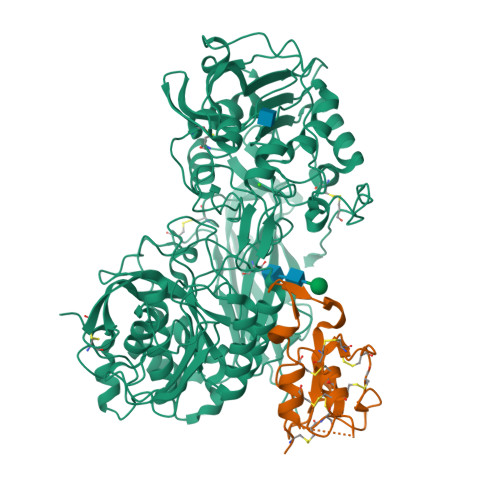
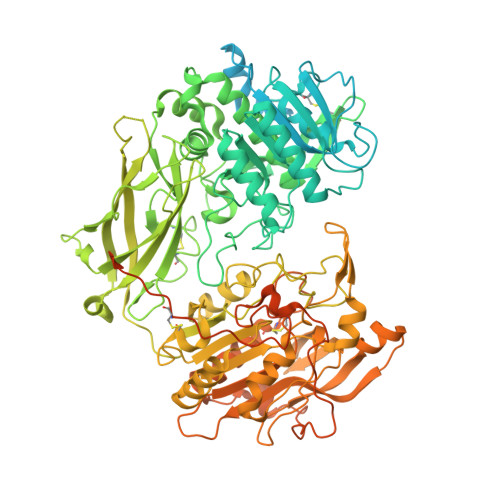
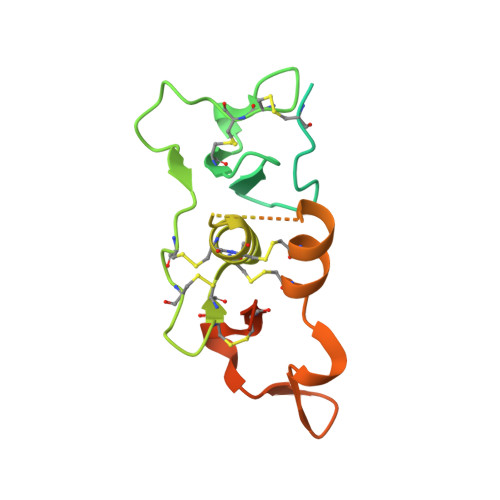
SPRING licenses S1P-mediated cleavage of SREBP2 by displacing an inhibitory pro-domain.
Hendrix, S., Dartigue, V., Hall, H., Bawaria, S., Kingma, J., Bajaj, B., Zelcer, N., Kober, D.L.(2024) Nat Commun 15: 5732-5732
- PubMed: 38977690
- DOI: https://doi.org/10.1038/s41467-024-50068-8
- Primary Citation of Related Structures:
8UW8, 8UWC - PubMed Abstract:
Site-one protease (S1P) conducts the first of two cleavage events in the Golgi to activate Sterol regulatory element binding proteins (SREBPs) and upregulate lipogenic transcription. S1P is also required for a wide array of additional signaling pathways. A zymogen serine protease, S1P matures through autoproteolysis of two pro-domains, with one cleavage event in the endoplasmic reticulum (ER) and the other in the Golgi. We recently identified the SREBP regulating gene, (SPRING), which enhances S1P maturation and is necessary for SREBP signaling. Here, we report the cryo-EM structures of S1P and S1P-SPRING at sub-2.5 Å resolution. SPRING activates S1P by dislodging its inhibitory pro-domain and stabilizing intra-domain contacts. Functionally, SPRING licenses S1P to cleave its cognate substrate, SREBP2. Our findings reveal an activation mechanism for S1P and provide insights into how spatial control of S1P activity underpins cholesterol homeostasis.
Organizational Affiliation:
Department of Medical Biochemistry, Amsterdam UMC, Amsterdam Cardiovascular Sciences and Gastroenterology and Metabolism, University of Amsterdam, Meibergdreef 9, 1105AZ, Amsterdam, the Netherlands.