Structure of CXCR2 bound to CXCL2 (CXCR2-CXCL2-Go Full map)
Sano, F.K., Saha, S., Sharma, S., Ganguly, M., Shihoya, W., Nureki, O., Shukla, A.K., Banerjee, R.To be published.
Experimental Data Snapshot
Starting Model: in silico
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Guanine nucleotide-binding protein G(o) subunit alpha | 240 | Homo sapiens | Mutation(s): 6 Gene Names: GNAO1 EC: 3.6.5 Membrane Entity: Yes | 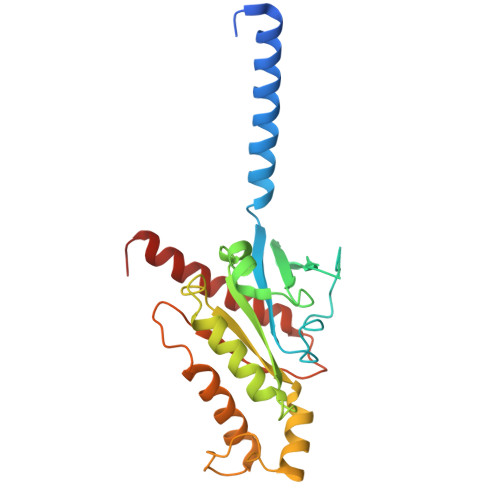 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P09471 (Homo sapiens) Explore P09471 Go to UniProtKB: P09471 | |||||
PHAROS: P09471 GTEx: ENSG00000087258 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P09471 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 | 350 | Homo sapiens | Mutation(s): 0 Gene Names: GNB1 | 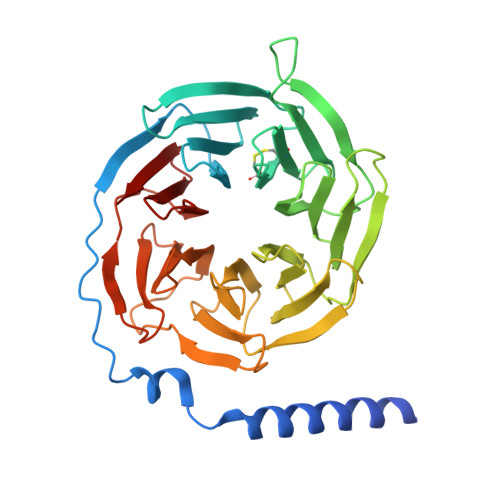 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P62873 (Homo sapiens) Explore P62873 Go to UniProtKB: P62873 | |||||
PHAROS: P62873 GTEx: ENSG00000078369 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62873 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 | C [auth G] | 71 | Homo sapiens | Mutation(s): 0 Gene Names: GNG2 Membrane Entity: Yes | 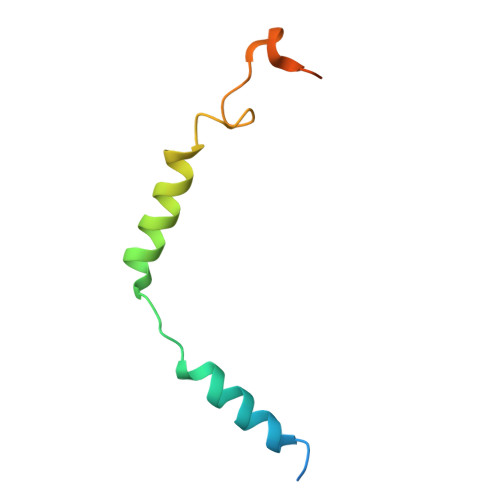 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P59768 (Homo sapiens) Explore P59768 Go to UniProtKB: P59768 | |||||
PHAROS: P59768 GTEx: ENSG00000186469 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P59768 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Antibody fragment ScFv16 | D [auth S] | 248 | Mus musculus | Mutation(s): 0 | 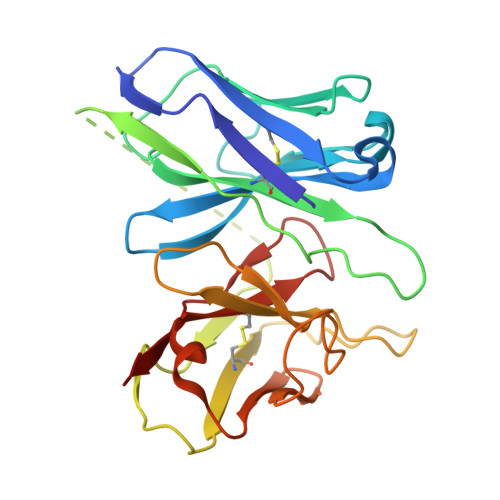 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| C-X-C chemokine receptor type 2 | E [auth R] | 416 | Homo sapiens | Mutation(s): 0 Gene Names: CXCR2 Membrane Entity: Yes | 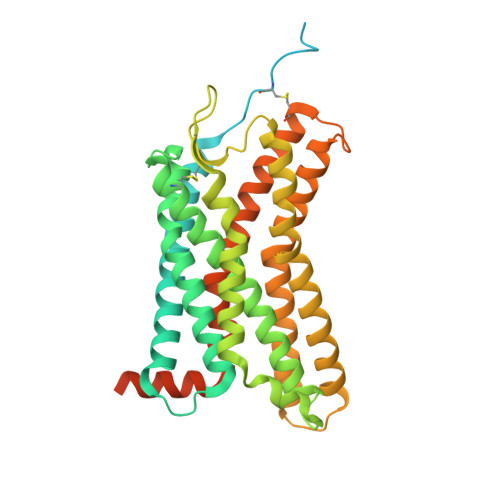 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P25025 (Homo sapiens) Explore P25025 Go to UniProtKB: P25025 | |||||
PHAROS: P25025 GTEx: ENSG00000180871 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P25025 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| C-X-C motif chemokine 2 | F [auth D], G [auth E] | 73 | Homo sapiens | Mutation(s): 0 Gene Names: CXCL2 Membrane Entity: Yes | 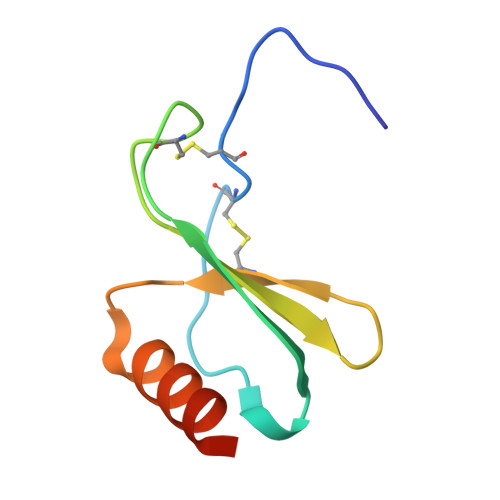 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P19875 (Homo sapiens) Explore P19875 Go to UniProtKB: P19875 | |||||
PHAROS: P19875 GTEx: ENSG00000081041 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P19875 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| RECONSTRUCTION | cryoSPARC | 4.4 |
| RECONSTRUCTION | RELION | 4 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Science and Engineering Research Board (SERB) | India | IPA/2020/000405 |