Fluorination Influences the Bioisostery of Myo-Inositol Pyrophosphate Analogs.
Hostachy, S., Wang, H., Zong, G., Franke, K., Riley, A.M., Schmieder, P., Potter, B.V.L., Shears, S.B., Fiedler, D.(2023) Chemistry 29: e202302426-e202302426
- PubMed: 37773020
- DOI: https://doi.org/10.1002/chem.202302426
- Primary Citation of Related Structures:
8G9C, 8G9D, 8G9E - PubMed Abstract:
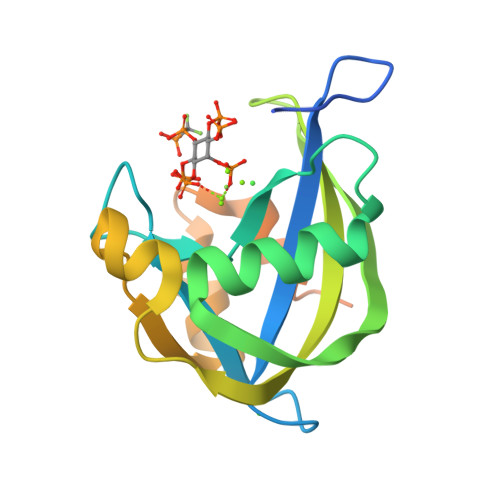
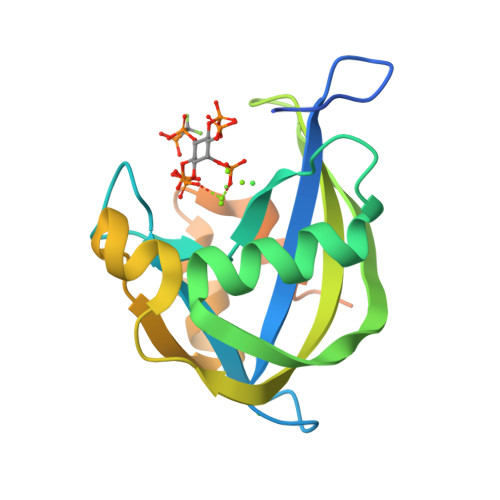
Inositol pyrophosphates (PP-IPs) are densely phosphorylated messenger molecules involved in numerous biological processes. PP-IPs contain one or two pyrophosphate group(s) attached to a phosphorylated myo-inositol ring. 5PP-IP 5 is the most abundant PP-IP in human cells. To investigate the function and regulation by PP-IPs in biological contexts, metabolically stable analogs have been developed. Here, we report the synthesis of a new fluorinated phosphoramidite reagent and its application for the synthesis of a difluoromethylene bisphosphonate analog of 5PP-IP 5 . Subsequently, the properties of all currently reported analogs were benchmarked using a number of biophysical and biochemical methods, including co-crystallization, ITC, kinase activity assays and chromatography. Together, the results showcase how small structural alterations of the analogs can have notable effects on their properties in a biochemical setting and will guide in the choice of the most suitable analog(s) for future investigations.
Organizational Affiliation:
Leibniz-Forschungsinstitut für Molekulare Pharmakologie (FMP), Robert-Rössle-Straße 10, 13125, Berlin, Germany.