Structural mechanism of CB1R binding to peripheral and biased inverse agonists
Kumari, P., Szabolcs, D., Enos, M.D., Ramesh, K., Lim, D., Hassan, S.A., Kunos, G., Iyer, M.R., Rosenbaum, D.M.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cannabinoid receptor 1,Glycogen synthase | A [auth R] | 535 | Homo sapiens, Pyrococcus abyssi GE5 | Mutation(s): 0 Gene Names: CNR1, CNR, PAB2292 | 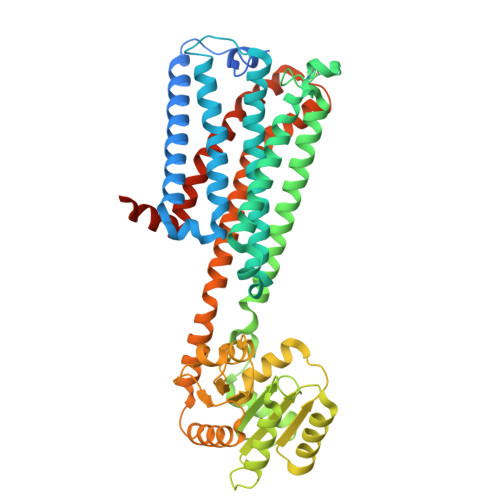 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9V2J8 (Pyrococcus abyssi (strain GE5 / Orsay)) Explore Q9V2J8 Go to UniProtKB: Q9V2J8 | |||||
Find proteins for P21554 (Homo sapiens) Explore P21554 Go to UniProtKB: P21554 | |||||
PHAROS: P21554 GTEx: ENSG00000118432 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Groups | Q9V2J8P21554 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| CNb36 | B [auth N] | 128 | Lama glama | Mutation(s): 0 | 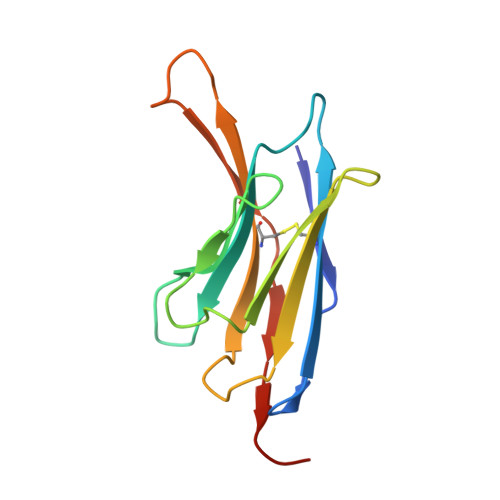 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 7DY (Subject of Investigation/LOI) Query on 7DY | C [auth R] | N-[(2S,3S)-4-(4-chlorophenyl)-3-(3-cyanophenyl)butan-2-yl]-2-methyl-2-{[5-(trifluoromethyl)pyridin-2-yl]oxy}propanamide C27 H25 Cl F3 N3 O2 QLYKJCMUNUWAGO-GAJHUEQPSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | Coot | |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | GM116387 |