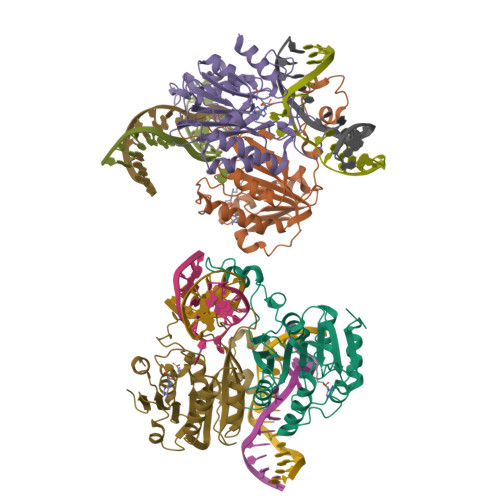
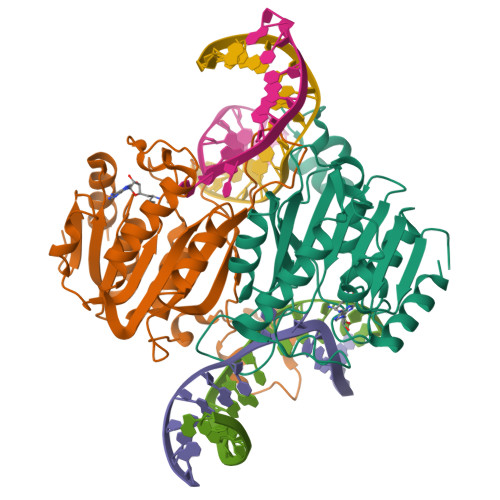
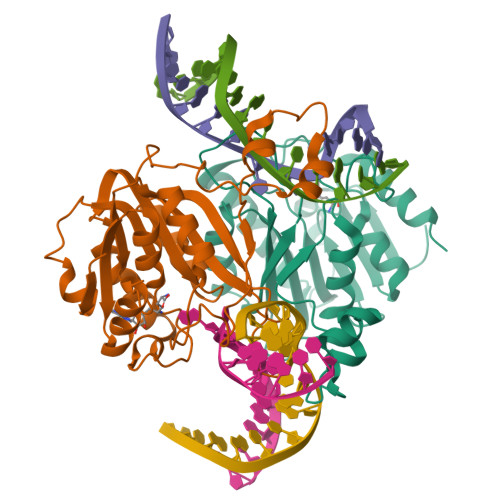
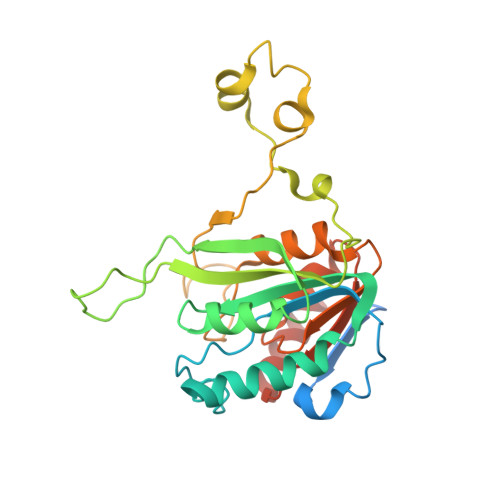
Burkholderia cenocepacia epigenetic regulator M.BceJIV simultaneously engages two DNA recognition sequences for methylation.
Quintana-Feliciano, R., Kottur, J., Ni, M., Ghosh, R., Salas-Estrada, L., Ahlsen, G., Rechkoblit, O., Shapiro, L., Filizola, M., Fang, G., Aggarwal, A.K.(2024) Nat Commun 15: 7839-7839
- PubMed: 39244607
- DOI: https://doi.org/10.1038/s41467-024-52130-x
- Primary Citation of Related Structures:
8URK, 9C3S, 9C3T, 9C3U - PubMed Abstract:
Burkholderia cenocepacia is an opportunistic and infective bacterium containing an orphan DNA methyltransferase called M.BceJIV with roles in regulating gene expression and motility of the bacterium. M.BceJIV recognizes a GTWWAC motif (where W can be an adenine or a thymine) and methylates N6 of the adenine at the fifth base position. Here, we present crystal structures of M.BceJIV/DNA/sinefungin ternary complex and allied biochemical, computational, and thermodynamic analyses. Remarkably, the structures show not one, but two DNA substrates bound to the M.BceJIV dimer, with each monomer contributing to the recognition of two recognition sequences. We also show that methylation at the two recognition sequences occurs independently, and that the GTWWAC motifs are enriched in intergenic regions in the genomes of B. cenocepacia strains. We further computationally assess the interactions underlying the affinities of different ligands (SAM, SAH, and sinefungin) for M.BceJIV, as a step towards developing selective inhibitors for limiting B. cenocepacia infection.
Organizational Affiliation:
Department of Pharmacological Sciences, Icahn School of Medicine at Mount Sinai, New York, New York, USA.